Team:HKU-HKBU/Protocol
From 2009.igem.org
PROTOCOL-1 β-galactosidase assay
1. Sample treatment
a) Harvest the culture by centrifuge (13krpm*1.5min) followed by washing with 1ml PBS twice
b) Resuspend the pellet with PBS and protease inhibitor cocktail.
c) Sonication for (5.5seconds+1second pulse)*10 minutes
d) Centrifuge (13krpm * 1min)
2. ONPG enzymatic reaction
a) Aliquot 200 microliter of treated sample into the 96 wells plate
b) Add 40 microliter of ONPG(4mg/ml in ddw) into each well
c) Read the OD420 every 10minutes for 5 minutes at 37’C on the spectrometer
3. Trouble shooting
a) Raw reading beyond most accurate region of o.2~0.4
b) Loss of activity due to high temperature or malfunction of the protease inhibitor
c) Incomplete cell lysis because too short sonication time or incomplete resuspension of the pellet
d) The temperature for the enzymatic reaction should be 37’C, so the spectrometer should be prewarmed.
<Note>
Must keep the sample at low temperature to avoid the loss of enzymatic activity.
PROTOCOL-2 Western blot
-
Buffer preparation
-
Rususpension buffer: 1ml stock + 0.1ml protease inhibitor (10x)
-
Loading buffer: 1ml stock + 0.2 ml DTT + Phenol Blue
-
-
Sample treatment
-
centrifuge the culture into pellet (13k rpm 10min)
-
Resuspend the pellet with 5xVolume(pellet) Resuspension buffer
-
Add 5xVolume(pellet) loading buffer
-
Boil the resuspend for 10 minutes at 100’C
-
Centrifuge for 1 min at 13krpm
-
-
SDS-PAGE
-
15% separation gel
-
5% stacking gel
-
Load the sample and run under the constant voltage of 100V
-
-
Western blotting
-
Transfer from gel to membrane
-
Assemble "sandwich" Transblot.
-
Prewet the sponges, filter papers (slightly bigger than gel) in 1x Blotting buffer.
-
Transfer for 1 hr at 1 amp at 4°C on a stir plate. Bigger proteins might take longer to transfer. For the Mini-Transblot, it's 100 V for 1 hr with the cold pack and prechilled buffer. When finished, immerse membrane in Blocking buffer and block overnight.
-
-
Hybridization with antibodies
-
Incubate with primary antibody diluted in Blocking buffer for 60 min at room temp.
-
Wash 3 x 10 min with 0.05% Tween 20 in PBS.
-
Incubate with secondary antibody diluted in PBS for 45 min at room temp.
-
Wash 3 x 10 min with 0.05% Tween 20 in PBS.
-
Detect with SUPER SIGNAL WEST PICO Kit (1ml luminol solution + 1ml stable peroxide solution
-
-
Result analysis
-
The size of the tagged protein can be determined by the marker (protein ladder)
-
The amount of the protein can be estimated by the brightness of the band, or accurately analysed by the software.
-
-
Trouble shooting
-
Unspecific binding : 1st antibody-membrane and irrelevant protein to 1st antibody
-
Film over-exposure or lack of exposure
-
PROTOCOL-3 Recombineering
The procedure used was similar to that described by Watt et al [1]. About 500 μl from overnight cultures [5 ml L medium (containing antibiotic where applicable) inoculated from single colonies, grown at 32°C for 18 h] was expanded into 50 ml of L medium in a 250 ml Erlenmeyer flask, and incubated at 32°C for 2 h (until OD600 of ca. 0.4–0.6). Flasks were transferred to a shaking water bath at 42°C and incubated for 14–15 min, before cooling to 0°C as rapidly as possible in iced water. After 15–20 min, cells were harvested by centrifugation at 0°C (4000 g, 9 min). Cell pellets were carefully washed three times with sterilized ice-cold water (2 × 50 ml, then 1 × 1.5 ml) then re-suspended in 100–200 μl of ice-cold water. Competent cells (50 μl) were transformed with 50–200 ng of (gel purified) linear dsDNA targeting cassette using a BioRad electroporator (1.8 kV, 25 mF, 200 W). The L medium (1 ml) was added to the transformed cell mixture, which was incubated at 32°C, for 2–2? h. Cells were collected by centrifugation, ca. 900 μl of supernatant media was discarded, and then the resuspended cells were plated onto LB agar containing the appropriate antibiotic to select for resistant colonies.
PROTOCOL-4 Most Probable Number method (MPN)
Preparation:
-
A original undiluted cell culture sample (> 1ml)
-
Distilled water
-
Vortex**
-
OD600 reader
-
Fresh medium (liquid or solid)
Steps:
-
Measure and record the OD600 value of the original undiluted sample.
-
Shake the sample thoroughly and draw 100μl into a new tube which contains 900μl water. Mark the new tube with 1.
-
Repeat step 2 for several times according to the OD600 value which have been recorded. Every time dilute the previous sample 10 times and mark the new sample in an ascending order, i.e. 2, 3, 4… (Since it is commonly believed that there are around 109 cells in 1ml sample with OD600 measured 1.00, if this sample is used, loop for 9 or 10 times is needed)
-
Up to this step, a series of diluted samples should be obtained. Every time draw 100μl from the sample and drop it on a plate. For every sample, repeat this procedure for 5 separate plates.
-
Incubate the plates over night, at 37℃.
-
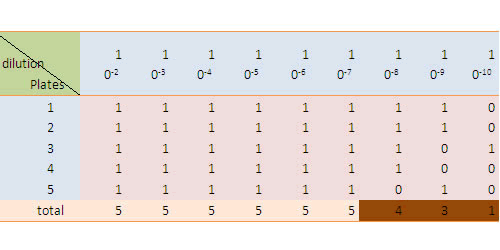
Record the outcome in the following format, in which 1 represents a bacteria growth is observed while 0 means not.
-
Check up the http://www.jlindquist.net/generalmicro/102dil3a.html MPN table for the final result.
To see our result, please refer to the RESULT part of our wiki.
PROTOCOL-5 Bradford assay
1. Bacteria culturing
A 50 mL culture of EColi. was grown in 2% LB media overnight (16 hours) in 37’C shaker. A fresh 50 mL LB culture was inoculated with 0.2 mL of these cells and grown for 5 hours to an OD600 between 0.100 and 3.000. The cells were collected with centrifugation (Beckman, 13,000rpm, 2min, rt’C), washed twice with chilled 200 λ PBS Buffer, and re-suspended in 1mL of chilled PBS Buffer.
2. Protein quantization
Sonication Settings:
1,000 μl aliquots of re-suspended bacteria were transferred to 1.4 mL polyethylene tubes tubes were subjugated to sonication times of (10”+5”)*10cycles seconds at 40% power (≈4.8 watts/pin). The samples were then centrifuged at 13,000 rpm for 2 minutes to pellet debris. The supernatant containing solubilized proteins was collected for analysis.
Or B-Per protein extraction kit(recommended):
1000 μL aliquots of re-suspended bacteria were centrifuged at 13k rpm for 10 min. to pellet cells. The cells were resuspended in 200 μL of B-Per Reagent (Pierce, Rockford, IL) and vortexed for 1 minute corresponding to the Pierce protocol. 800 μl of PBS buffer was added to the solution and the solution centrifuged 10min 13k rpm to pellet debris and the supernatant collected for analysis
Protein Assay:
The amount of protein released after each sonication time was qualitatively determined by use of Bradford Reagent. An aliquot of 10 μl of each sample was mixed with 200 μl of 1:5Diluted Bradford Reagent and the absorption at 595 nm recorded (CARY300 Bio UV-Visible spectrometer) after 10~30 minutes of mixing time.
PROTOCOL-6 Casting agarose gels
-
Assemble casting trays, gel box and gel comb, choose the comb of appropriate width, size, and number into niche at end of the tray.
-
agarose solution(1% gel):0.4g agarose + 40ml 1X TAE for small gels or 1g agarose + 100ml 1X TAE for large gels.
-
add agarose solution in a 500ml beaker.
-
Microwave bottle for 5min
-
Remove from microwave and let cool
-
Once gel is cooled so that it can be touched comfortably with your gloved hand, add 4μL(for small gel) or 10 μL(for large gels) EB dye.
-
Pour gel into casting trays.
PROTOCOL-7 Running agarose gels
-
Place the gel in gel box.
-
Add enough 1X TAE to fill the reservoirs at both ends of the gel box and cover the surface of the gel--the gel should be immersed.
-
Mix6Х loading dye with samples: Typically, use 1 μL loading dye per 5 μL of sample.
-
Load samples left to right.
-
Place gel box cover on gel box such that your samples will run towards the positive, red electrode. Make sure that the cables from the cover are connected to the power supply correctly.
-
Turn on the power supply and run your gel at ~120 V for 30 mins.
-
Verify that bubbles are rising from the electrodes once you start your gel to ensure your gel is running properly.
Marker standard
We use 1kb plus DNA ladder.
PROTOCOL-8 Quick gel purifcation
according to QIAprep kit protocol
-
Agarose gel electrophoresis, cut the DNA band as thin as possible.Then put it into a clear EP tube. weighing,if the weight is 100mg,the volume can be considered as 100uL,add QG beffer,put into 55℃ water bath until complently merge, it takes about 10 minutes
-
cool the tube to room temperature
-
Add the solution into the collection tube, standing for 1 minute,13,000rpm for 1 min, discard the flow-through
-
Add 650 μl WB,13,000rpm for 1 min, discard the flow-through
-
Again
-
13,000rpm for 2 min, discard the flow-through
-
Put the collection tube into a clean EP tube, Add 30 μl ddH2O in the center of the collection tube, standing for 5min
-
13,000rpm for 2 min.
-
store the DNA solution at -20℃
PROTOCOL-9 PCR product purifcation
according to QIAprep kit protocol
-
Take 100uL PCR product solution, add 5 times volume Binding Buffer,mix, add into spin column, standing for 2 min, 13000rpm for 1 min,discard the flow-through.
-
Add 650 mL buffer WB ,13000 rpm for 1min,discard the supernatant
-
Again
-
13000rpm for 2 min to wipe out the remaining WB
-
Put the collection tube into a clean EP tube, Add 50uL 60 ℃ddH2O in the center of the collection tube, standing for 15min
-
13000rpm for 12min.
-
store the DNA solution at -20℃
PROTOCOL-10 Count: Preparation of Competent Cells for electro transformation
Materials
-
Media
-
LB(Both liquid media and media containing agar. Add certain antibiotic if it is necessary.);
-
Buffers and Solutions
10% Glycerin.
-
Special Equipments
-
EP tubes(1.5mL), micropipette tips, centrifugation bottles(polypropylene tubes, 50mL), graduated flask(250mL*2, 5mL*1), plates and test tubes.
Protocol
-
Sterilization
-
Including all materials in part2.
-
Caution (Remind): use some special marks to distinguish the sterilized materials from the unsterilized ones.
-
Preparation after sterilization
-
Chill the 10%Glycerin to 4 centigrade degree
-
Decant LB containing agar into the plates.(If antibiotics are necessary, be sure that they are added when the media temperature is below 60 centigrade degree.)
-
Streak the prepared strains onto the agar plates. Then incubate it at 37℃ for 10-16 hours.
-
overnight preculture
-
take? 0.5mL overnight culture to 50mL LB bottle.
-
37℃ shaking 100~120 min to O.D. 600=0.45~0.6
-
on ice for 30min
-
4000rpm 7min at 4℃
-
Add origin volume 10% glycerol,suspend softly.
-
4000rpm7 min at 4℃
-
Add origin volume 10% glycerol,suspend softly.
-
Add origin volume 1/10 10% glycerol, suspend softly.
-
4000rpm7 min at 4℃
-
Add origin volume 1/100 10% glycerol, suspend softly, store at -80℃.
PROTOCOL-11 Electro tranformation
-
Hold competent cells (from -80℃ refrigerator) on ice.
-
Gently mix ligation product (1-5 μL) with cells.
-
transfer the cell/DNA mix into an electroporation cuvette
Note: the gene pulser should already be set properly
- time constant = 4.5 - 5.0 ms
- resistance = 200 W
- capacitance = 25 mFD
for 0.1 cm gap cuvettes, set the volts to 1.7 kV -
pulse the cells once; the voltage display blinks, and the gene pulser beeps
-
quickly transfer 37 ℃ SOC to cuvette, mix by gently pipetting up and down, and transfer SOC/cells back to culture tube
-
Bath in 37℃ for 60~90 min.?
-
Separate cells on petri-dishes, and cultivate them in 37℃ for 12 hour.
PROTOCOL-12 Ligation
We use new England Biolabs T4 DNA ligase
-
Choose reaction volume: 5-10 μL
-
Mix proper proportion (usually 3~5:1) of DNA fragment and vector.
-
Add 10×(meaning 1/10 of final volume) ligase buffer.
-
Add 0.5 μL ligase per 10 μL final volume.
-
Bath in 16℃ water for 12 hour,
-
Begin http://www.openwetware.org/wiki/IGEM:Peking/2007/Transformation transformation.
PROTOCOL-13 DNA extraction
Use QIAGEN kit
PROTOCOL-14 Polymerase Chain Reaction
Choose enzymes
1.We use rTaq in testing but not in cloning genes.
2.Ex Taq and LA Taq are modified Taq by Takara, they have 3’->5’exonuclease activity and a relatively high fidelity.
3.Generally speaking, genes shorter than 1000bp can be cloned by Ex Taq. Genes shorter than 1.5kb can be cloned by LA Taq.
4.Taq can add A at the end of each fragment, so their PCR product can direcly link to T plasmid.
PCR system
Normal Taq polymerase reaction component has a template of 1ng~1ug, dNTP 200uM each,primer 50pmol each,polymerase 1U,buffer and water。Template can be plasmid,colony and genomic DNA.
Taq series polymerase reaction components
Colony PCR
Template:Bacterial Colony
10×PCR rTaqbuffer 2uL
10×dNTP 1.6uL
Primer-F 0.1uL
Primer-R 0.1uL
rTaq 0.2uL
ddH2O 16uL
20uL system
Cloning
Template 1uL (above 10ngDNA)
10×PCR buffer(Ex/LA) 5uL
10×dNTP 4uL
Primer-F 1uL
Primer-R 1uL
Ex/LA Taq 0.25uL
ddH2O 37.75uL
50uL system
reaction condition
1.95℃ 5min Taq enzyme activation by heat
2.95℃ 30s DNA denaturing
3.Tm-5~10℃ 30s Tm is annealing temperature, with a range of 45~60℃
4. 72℃ ETs ET is elongation time,1kb/min
28cycles
5. 72℃ 10min add A at the end of each fragment
PROTOCOL-15 Plasmid digestion
Digestion temperature
Most endonuclease has the optimal activity temperature of 37℃.
enzymatic system
Includs plasmid/PCR product, enzyme, buffer and water, sometimes with BSAin it. The system for enzymatic test should be no more than 20uL. The system for recruiting vectors and fragments should be 50-100uL.
DNA concentration in digestion system
The vector concentration should be no more than 1ug, PCR fragment concentration should be no less than 1ug.
Star activity
Some endonuclease will have a less specific substrate selection under certain conditions. It will cut different sequences from original recognitions. This is called star activity.
In order to reduce the star activity, glycero concentration should not be too high and DNA concentration should not be too low. In the storage of enzyme there is glycerol. So the maximal amount of enzyme added in a system should be 10%, especially for enzymes with star activities.
The influence of Methylation
Some endonuclease activity will be affected by methylation of DNA.
There is a methylation form we can refer to.
Terminal digestion of PCR product
The terminal digestion always has a low efficiency. Some additional base pairs are always added at the end of PCR product during primer design in order to improve the digestion efficiency.
There is a form about PCR terminal digestion.
Inactivation
After the digestion, endonuclease must be inactivated before applied to ligation system. Different enzymes have different inactivation condition.
There is a form about inactivation conditions of all usual endonuclease.
Digestion Timing
1 hour enables a test digestion. If digestion is used for cutting vectors and PCR fragments, in order to digest thoroughly, about 16 hours is a relatively good choice. However, some endonuclease used in the digestion system inactivates quickly. So we need add new enzymes at intervals during digestion.
Refer to the form of different endonuclease about their remaining activity after different periods of time.
Reference
- Watt RM, Wang J, Leong M, Kung HF, Cheah KS, Liu D, Danchin A, Huang JD. Visualizing the proteome of Escherichia coli: an efficient and versatile method for labeling chromosomal coding DNA sequences (CDSs) with fluorescent protein genes. Nucleic Acids Res. 2007, 35(6):e37.
 "
"