Team:Heidelberg/Notebook natural promoters
From 2009.igem.org
(Difference between revisions)
(→8-31-2009) |
|||
| (54 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
| + | {{Template_HD_3}} | ||
| + | <html> | ||
| + | <style = "text/css"> | ||
| + | #body {margin: 0} | ||
| + | </style> | ||
| + | <body id="project"></body></html> | ||
| - | { | + | {| valign="top" border="0" |
| - | + | |width="900px" style="padding: 0 15px 15px 20px; background-color:#ede8e2"| | |
| - | + | ||
| - | | | + | |
| - | |width=" | + | |
__NOTOC__ | __NOTOC__ | ||
| - | |||
=='''Natural Promoters'''== | =='''Natural Promoters'''== | ||
=='''Contents'''== | =='''Contents'''== | ||
| - | {| class="wikitable centered" border="2" rules="rows" width=" | + | {| class="wikitable centered" border="2" rules="rows" width="900px" style="border-color:white;" |
|- | |- | ||
! Week !! colspan="7" |Days | ! Week !! colspan="7" |Days | ||
| Line 39: | Line 41: | ||
|style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#8-27-2009|8-27-2009]] | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#8-27-2009|8-27-2009]] | ||
|style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#8-28-2009|8-28-2009]] | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#8-28-2009|8-28-2009]] | ||
| + | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#8-29-2009|8-29-2009]] | ||
|style="text-align:center"| - | |style="text-align:center"| - | ||
| + | |- | ||
| + | |style="text-align:center"| 36 | ||
| + | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#8-31-2009|8-31-2009]] | ||
| + | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#9-01-2009|9-01-2009]] | ||
| + | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#9-02-2009|9-02-2009]] | ||
| + | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#9-03-2009|9-03-2009]] | ||
| + | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#9-04-2009|9-04-2009]] | ||
|style="text-align:center"| - | |style="text-align:center"| - | ||
| + | |style="text-align:center"| - | ||
| + | |- | ||
| + | |style="text-align:center"| 37 | ||
| + | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#9-07-2009|9-07-2009]] | ||
| + | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#9-08-2009|9-08-2009]] | ||
| + | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#9-09-2009|9-09-2009]] | ||
| + | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#9-10-2009|9-10-2009]] | ||
| + | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#9-11-2009|9-11-2009]] | ||
| + | |style="text-align:center"| - | ||
| + | |style="text-align:center"| - | ||
| + | |- | ||
| + | |style="text-align:center"| 38 | ||
| + | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#9-14-2009|9-14-2009]] | ||
| + | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#9-15-2009|9-15-2009]] | ||
| + | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#9-16-2009|9-16-2009]] | ||
| + | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#9-17-2009|9-17-2009]] | ||
| + | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#9-18-2009|9-18-2009]] | ||
| + | |style="text-align:center"| - | ||
| + | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#9-20-2009|9-20-2009]] | ||
| + | |- | ||
| + | |style="text-align:center"| 39 | ||
| + | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#9-21-2009|9-21-2009]] | ||
| + | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#9-22-2009|9-22-2009]] | ||
| + | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#9-23-2009|9-23-2009]] | ||
| + | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#9-24-2009|9-24-2009]] | ||
| + | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#9-25-2009|9-25-2009]] | ||
| + | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#9-26-2009|9-26-2009]] | ||
| + | |style="text-align:center"| - | ||
| + | |- | ||
| + | |style="text-align:center"| 40 | ||
| + | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#9-28-2009|9-28-2009]] | ||
| + | |style="text-align:center"| [[Team:Heidelberg/Notebook_natural_promoters#9-29-2009|9-29-2009]] | ||
| + | |style="text-align:center"| - | ||
| + | |style="text-align:center"| - | ||
| + | |style="text-align:center"| - | ||
| + | |style="text-align:center"| - | ||
| + | |style="text-align:center"| - | ||
| + | |- | ||
|} | |} | ||
| - | ''' plasmid p31 = plasmid pSMB_MEASURE ''' | + | * ''' plasmid p31 = plasmid pSMB_MEASURE ''' |
== 8-11-2009 == | == 8-11-2009 == | ||
| Line 50: | Line 98: | ||
{| class="wikitable centered" style="margin: 1em auto 1em auto" | {| class="wikitable centered" style="margin: 1em auto 1em auto" | ||
|- style="background-color:#99cccc;" | |- style="background-color:#99cccc;" | ||
| - | |style="text-align:center"|Plasmids || Source | + | |style="text-align:center"|Plasmids ||style="text-align:center"| Source |
|- style="background-color:#9BDDFF;" | |- style="background-color:#9BDDFF;" | ||
| - | |style="text-align:left"| Plasmid with NFAT responsive promoter|| Oakes | + | |style="text-align:left"| Plasmid with NFAT responsive promoter|| Dr. C. Oakes, Heidelberg (DKFZ) |
|- style="background-color:#9BDDFF;" | |- style="background-color:#9BDDFF;" | ||
| - | |style="text-align:left"| Plasmid with NFkB responsive promoter || Oakes | + | |style="text-align:left"| Plasmid with NFkB responsive promoter || Dr. C. Oakes, Heidelberg (DKFZ) |
|- style="background-color:#9BDDFF;" | |- style="background-color:#9BDDFF;" | ||
| - | |style="text-align:left"| pJC6-GL3 with c-Jun promoter || Addgene | + | |style="text-align:left"| pJC6-GL3 with c-Jun promoter || Addgene, Cambridge |
|- style="background-color:#9BDDFF;" | |- style="background-color:#9BDDFF;" | ||
| - | |style="text-align:left"| SBE4-Luc with Smad binding element promoter || Addgene | + | |style="text-align:left"| SBE4-Luc with Smad binding element promoter || Addgene,Cambridge |
|- style="background-color:#9BDDFF;" | |- style="background-color:#9BDDFF;" | ||
| - | |style="text-align:left"| pGL3-RARE-Luciferase with Retinoic Acid Receptor Response Element || Addgene | + | |style="text-align:left"| pGL3-RARE-Luciferase with Retinoic Acid Receptor Response Element || Addgene, Cambridge |
|- style="background-color:#9BDDFF;" | |- style="background-color:#9BDDFF;" | ||
| - | |style="text-align:left"| pGL3-NFAT-Luciferase with NFAT responsive promoter || Addgene | + | |style="text-align:left"| pGL3-NFAT-Luciferase with NFAT responsive promoter || Addgene, Cambridge |
|- style="background-color:#9BDDFF;" | |- style="background-color:#9BDDFF;" | ||
| - | |style="text-align:left"| PUMA-Frag1-Luc eith PUMA promoter|| Addgene | + | |style="text-align:left"| PUMA-Frag1-Luc eith PUMA promoter|| Addgene, Cambridge |
|- style="background-color:#9BDDFF;" | |- style="background-color:#9BDDFF;" | ||
| - | |style="text-align:left"| pLDLR-Luc with LDL receptor promoter || Addgene | + | |style="text-align:left"| pLDLR-Luc with LDL receptor promoter || Addgene, Cambridge |
|- style="background-color:#9BDDFF;" | |- style="background-color:#9BDDFF;" | ||
| - | |style="text-align:left"| pHMGCS-Luc with HMG CoA synthase promoter|| Addgene | + | |style="text-align:left"| pHMGCS-Luc with HMG CoA synthase promoter|| Addgene, Cambridge |
|- style="background-color:#9BDDFF;" | |- style="background-color:#9BDDFF;" | ||
| - | |style="text-align:left"| p52 plasmid with HSP70 promoter || | + | |style="text-align:left"| p52 plasmid with HSP70 promoter || M. Schmidt and F. Hoppe, Würzburg |
|} | |} | ||
== 8-18-2009 == | == 8-18-2009 == | ||
| - | * Start | + | * Start of natural promoter project |
| - | * The two promoters, Apolipoprotein A-IV (APOAIV) and Cytochrome P450 1A1 (CYP1A1) promoter, were amplified by PCR | + | * The two promoters, Apolipoprotein A-IV (APOAIV) and Cytochrome P450 1A1 (CYP1A1) promoter, were amplified by PCR from human genomic DNA using primers with BBB_standard. |
== 8-19-2009 == | == 8-19-2009 == | ||
* APOAIV promoter PCR product was digested with NheI and SpeI at 37°C for one hour. | * APOAIV promoter PCR product was digested with NheI and SpeI at 37°C for one hour. | ||
| - | * The backbone plasmid (p31; ~5000 bp) was also digested with the same restriction enzymes (at 37°C, 1 hour) and it was subsequent digested with SAP (at 37°C, 1 hour). | + | * The backbone plasmid (p31; ~5000 bp) was also digested with the same restriction enzymes (at 37°C, 1 hour) and it was subsequent digested with SAP (at 37°C, 1 hour). The insert of this plasmid p31 is the promoter JeT, which will be cut out after this digest. |
== 8-20-2009 == | == 8-20-2009 == | ||
| Line 89: | Line 137: | ||
* Picked eight colonies of p31/APOAIV plates and did a mini-preparation (miniprep) | * Picked eight colonies of p31/APOAIV plates and did a mini-preparation (miniprep) | ||
* Test digest of the eight miniprep's using the restriction enzymes SpeI and NheI (at 37°C, 1 hour) | * Test digest of the eight miniprep's using the restriction enzymes SpeI and NheI (at 37°C, 1 hour) | ||
| - | |||
* There is an appropriate band for the APOAIV promoter (~4000bp length). | * There is an appropriate band for the APOAIV promoter (~4000bp length). | ||
* Miniprep of colony p31/ApoAIV #1, p31/ApoAIV #2 and p31/ApoAIV #3 | * Miniprep of colony p31/ApoAIV #1, p31/ApoAIV #2 and p31/ApoAIV #3 | ||
== 8-24-2009 == | == 8-24-2009 == | ||
| - | * The plasmids p31/APOAIV #3 and p31/APOAIV #1 will be sequenced | + | * The plasmids p31/APOAIV #3 and p31/APOAIV #1 will be sequenced. |
== 8-25-2009 == | == 8-25-2009 == | ||
| Line 101: | Line 148: | ||
== 8-26-2009 == | == 8-26-2009 == | ||
* Troubleshooting PCR for CYP1A1 and APOAIV using Taq mastermix and Phu with freshly mixed ingredients. We also used various MgCl2 concentrations (1, 2, 3, 4 ul of 25 mM MgCl2 per 50ul reaction) and performed the reaction with and without the addition of DMSO. | * Troubleshooting PCR for CYP1A1 and APOAIV using Taq mastermix and Phu with freshly mixed ingredients. We also used various MgCl2 concentrations (1, 2, 3, 4 ul of 25 mM MgCl2 per 50ul reaction) and performed the reaction with and without the addition of DMSO. | ||
| - | * Analytic gel below shows CYP1A1 with Taq -/+ DMSO in the first two lanes with bands at the expected height of around 1200 bp. Lanes 1-10 are different CYP1A1 reactions, lanes 11-20 are APOAIV reactions. APOAIV shows only nonspecific bands, has to be further optimized using gradient PCR or new primers. | + | * Analytic gel below shows CYP1A1 with Taq -/+ DMSO in the first two lanes with bands at the expected height of around 1200 bp. Lanes 1-10 are different CYP1A1 reactions, lanes 11-20 are APOAIV reactions (Fig. 1). APOAIV shows only nonspecific bands, has to be further optimized using gradient PCR or new primers. |
| - | '''' | + | [[image:HD09_Trouble_CYP_APO.png|center|600px|thumb|'''Figure 1: 1% agarose gel electrophoresis of the PCR products of APOAIV and CYP1A1 promoter.'''The APOAIV and CYP1A1 promoter PCR products were analysed by agarose gel electrophoresis. Two reaction mixtures were used; one with Taq polymerase and the other one with Phu polymerase and with and without DMSO adding. Besides, the reaction mixtures of ''Phu'' have a varying concentration of MGCl2 (1-4 µl in a 50 µl reaction mixture). Additionally, a DNA ladder (100 - 10000 bp; Fermentas DNA Ladder Mix) is added into the gel for the validation of the DNA product length. The resulting CYP1A1 inserts should be 1229 bp long and APOAIV ~4200 bp. The "CYP1A1 ''Taq''" and "CYP1A1 ''Taq'' + DMSO" are the only products, which have the right length. All the other lanes show an unspecific PCR.]] |
== 8-27-2009 == | == 8-27-2009 == | ||
* Test digest of CYP1A1 insert (1.2 kb) with NheI (digest at position 614 bp) at 37°C for one hour. | * Test digest of CYP1A1 insert (1.2 kb) with NheI (digest at position 614 bp) at 37°C for one hour. | ||
| - | * PCR of NFkB responsive and NFAT responsive promoter plasmids to amplify promoters from Oakes plasmids p46 and p47. Amplified using PCR high annealing protocol: | + | * PCR of NFkB responsive and NFAT responsive promoter plasmids to amplify promoters from Oakes plasmids p46 and p47. Amplified using PCR high annealing protocol and primers with BBB_standard: |
{| class="wikitable centered" style="margin: 1em auto 1em auto" | {| class="wikitable centered" style="margin: 1em auto 1em auto" | ||
| Line 144: | Line 191: | ||
* Miniprep of p31 plasmid + NFAT/NFkB promoter. | * Miniprep of p31 plasmid + NFAT/NFkB promoter. | ||
* Ligation of CYP1A1 promoter and p31 plasmid at 22°C for 3 hours. | * Ligation of CYP1A1 promoter and p31 plasmid at 22°C for 3 hours. | ||
| - | * Test digest (EcoRI and PstI) of NFAT/NFkB promoter within the p31 plasmid ( | + | * Test digest (EcoRI and PstI) of NFAT/NFkB promoter within the p31 plasmid (Fig. 2)at 37°C for one hour. |
* NFAT#2 and NFAT#5 plasmids sent to sequencing. | * NFAT#2 and NFAT#5 plasmids sent to sequencing. | ||
| - | [[image:HD09_31.08._NFkB_NFAT.gif|center|600px|thumb|'''Figure | + | [[image:HD09_31.08._NFkB_NFAT.gif|center|600px|thumb|'''Figure 2: 1% agarose gel electrophoresis for a test digest of the NFkB/p31 plasmid and the NFAT/p31 plasmid.''' The NFkB/p31 and NFAT/p31 plasmids were digested with EcoRI and PstI at 37°C for one hour. The numbers (1,3,4,5 etc.) describe the plasmids of different bacterial colonies. Additionally, a DNA ladder (100 - |
10000 bp; Fermentas DNA Ladder Mix) is added into the gel for the validation of the DNA product length. The resulting NFAT insert should be 134 bp long and NFkB 179 bp. NFAT #2 and NFAT #5 seem to be successfully ligated plasmids, because there are small bands at ~ 150 bp. The lanes of NFkB #4 and NFkB #5 show a p31 plasmid band, which is 4919 bp long. But the NFkB inserts could not be recognized. Nevertheless, we will try to sequence them. All other lanes display definetly the wrong products.]] | 10000 bp; Fermentas DNA Ladder Mix) is added into the gel for the validation of the DNA product length. The resulting NFAT insert should be 134 bp long and NFkB 179 bp. NFAT #2 and NFAT #5 seem to be successfully ligated plasmids, because there are small bands at ~ 150 bp. The lanes of NFkB #4 and NFkB #5 show a p31 plasmid band, which is 4919 bp long. But the NFkB inserts could not be recognized. Nevertheless, we will try to sequence them. All other lanes display definetly the wrong products.]] | ||
| Line 164: | Line 211: | ||
== 9-04-2009 == | == 9-04-2009 == | ||
| - | * PCR with the plasmid p52 to isolate the HSP70 promoter | + | * PCR with the plasmid p52 to isolate the HSP70 promoter. Design primer with BBB standard, which were adjusted to the plasmid because of the lack of the p52 sequence. |
* PCR with the plasmid p46 to isolate the promoter with NFkB binding sites | * PCR with the plasmid p46 to isolate the promoter with NFkB binding sites | ||
* Digest of the plasmid p31 with the restriction enzymes, EcoRI and NheI (at 37°C, 1 hour) | * Digest of the plasmid p31 with the restriction enzymes, EcoRI and NheI (at 37°C, 1 hour) | ||
| Line 184: | Line 231: | ||
== 9-09-2009 == | == 9-09-2009 == | ||
* Gel electrophoresis image of the test digest: | * Gel electrophoresis image of the test digest: | ||
| - | + | * CYP1A1_promoter/p31 construct was included in three positive colonies (Fig. 3). Therefore, this construct is ready for mutagenesis PCR to remove the NheI restriction site at 614 bp. | |
| - | + | ||
| - | + | ||
| - | * CYP1A1_promoter/p31 construct was included in three positive colonies. Therefore, this construct is ready for mutagenesis PCR to remove the NheI restriction site at 614 bp | + | |
* The others have not turned out satisfactory. | * The others have not turned out satisfactory. | ||
| - | == 10 | + | [[image:HD09_CYP1A1.png|center|600px|thumb|'''Figure 3: 1% agarose gel electrophoresis for a test digest of the CYP1A1/p31 plasmid.''' The CYP1A1/p31 plasmids were digested with EcoRI and PstI at 37°C for one hour. The numbers (1,3,4,5 etc.) describe the plasmids of different bacterial colonies. Additionally, a DNA ladder (75 -20000 bp); Fermentas DNA 1kb Plus) is added into the gel for the validation of the DNA product length. The resulting CYP1A1 insert should be 1229 bp long. CYP1A1 #2, CYP1A1 #5 and CYP1A1 #8 seem to be successfully ligated plasmids, because there are small bands at ~ 1200 bp. All lanes show a p31 plasmid band, which is 4919 bp long. But other CYP1A1 inserts could not be recognized. All other lanes display definetly the wrong product (JeT, ~180 bp).]] |
| - | * The sequence of CYP1A1 #2 and #5 are right. | + | |
| + | == 9-10-09 == | ||
| + | * The sequence of CYP1A1 #2, #8 and #5 are right. | ||
* Plate out the Addgene plasmids: pJC6-GL3, SBE4-Luc, pGL3-RARE-Luc, pGL3-NFAT-Luc, PUMA-Frag1-Luc, pLDLR-Luc, pHMGCS-Luc | * Plate out the Addgene plasmids: pJC6-GL3, SBE4-Luc, pGL3-RARE-Luc, pGL3-NFAT-Luc, PUMA-Frag1-Luc, pLDLR-Luc, pHMGCS-Luc | ||
| - | == | + | == 9-11-09 == |
* Pick some colonies of the plates (10.09.09) and miniprep. | * Pick some colonies of the plates (10.09.09) and miniprep. | ||
| - | == | + | == 9-14-2009 == |
* PCR of HSP70, NFAT responsive and NFkB responsive promoters. | * PCR of HSP70, NFAT responsive and NFkB responsive promoters. | ||
* Digest of the PCR products by NheI and EcoRI at 37°C for one hour. | * Digest of the PCR products by NheI and EcoRI at 37°C for one hour. | ||
| Line 204: | Line 250: | ||
* Mutagenesis PCR of CYP1A1/p31 #5. | * Mutagenesis PCR of CYP1A1/p31 #5. | ||
| - | == | + | == 9-15-2009 == |
* PCR-Purification with HSP70, NFkB and NFAT | * PCR-Purification with HSP70, NFkB and NFAT | ||
* Gel-extraction of p31_BBB, which was digested by NheI and EcoRI at 37°C for one hour. | * Gel-extraction of p31_BBB, which was digested by NheI and EcoRI at 37°C for one hour. | ||
| Line 213: | Line 259: | ||
* Transformation of the three ligations (above: Hsp70/p31, NFAT/p31 and NFkB/p31) and CYP1A1-muta/p31 | * Transformation of the three ligations (above: Hsp70/p31, NFAT/p31 and NFkB/p31) and CYP1A1-muta/p31 | ||
| - | == | + | == 9-16-2009 == |
* Arrival of the sequencing results of the ordered Addgene plasmids. | * Arrival of the sequencing results of the ordered Addgene plasmids. | ||
* Design of primers for c-Jun promoter, LDL receptor promoter (LDLR), Retinoic Acid Receptor Response Element (RARE) and HMG CoA synthase promoter (HMG CoA) | * Design of primers for c-Jun promoter, LDL receptor promoter (LDLR), Retinoic Acid Receptor Response Element (RARE) and HMG CoA synthase promoter (HMG CoA) | ||
| Line 219: | Line 265: | ||
* p52 (with HSP70 insert) is amplified in bacteria (DH5alpha). | * p52 (with HSP70 insert) is amplified in bacteria (DH5alpha). | ||
* Miniprep of the following ligation : Hsp70 with p31_BBB, NFAT with p31_BBB and NFkB with p31_BBB | * Miniprep of the following ligation : Hsp70 with p31_BBB, NFAT with p31_BBB and NFkB with p31_BBB | ||
| - | + | * The PCR troubleshooting with NFAT and NFkB were analysed by a agarose gel and there were bright bands at ~300 bp, which are a little bit too long. Nevertheless, the inserts could be used for cloning. | |
| - | * The PCR troubleshooting with NFAT and NFkB were analysed by a agarose gel and there are | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | == 17 | + | == 9-17-2009 == |
| - | * | + | * Test digest of the ligations p52 (with HSP70 insert), NFAT and NFkB with EcoRI and PstI (37°C, 1 hour -> no right bands! |
* Ligation of NFAT and NFkB (of troubleshooting PCR) with p31 plasmid at 22°C for one hour. | * Ligation of NFAT and NFkB (of troubleshooting PCR) with p31 plasmid at 22°C for one hour. | ||
* No colonies of CYP1A1-muta -> try again the mutagenesis PCR (with special mutagenesis Kit) | * No colonies of CYP1A1-muta -> try again the mutagenesis PCR (with special mutagenesis Kit) | ||
| - | == | + | == 9-18-2009 == |
| - | * Design of primers for the HSP70 insert by analysis of p52 sequence | + | * Design of primers with BBB standard for the HSP70 insert by analysis of the p52 sequence. |
* Second mutagenesis PCR of CYP1A1 | * Second mutagenesis PCR of CYP1A1 | ||
* Transformation of NFAT and NFkB ligations | * Transformation of NFAT and NFkB ligations | ||
| - | == | + | == 9-20-2009 == |
* pick colonies of NFAT and NFkB | * pick colonies of NFAT and NFkB | ||
| - | == | + | == 9-21-2009 == |
| - | * PCR of RARE, LDLR, HMG CoA synthase and c-Jun promoter. There were two reaction mixture for each promoter. One with ''Taq'' polymerase and one with ''Phu'' polymerase. | + | * PCR of RARE, LDLR, HMG CoA synthase and c-Jun promoter using primer with BBB_standard. There were two reaction mixture for each promoter. One with ''Taq'' polymerase and one with ''Phu'' polymerase. |
* Miniprep of NFAT responsive and NFkB responsive promoters and test digest at 37°C for one hour -> no right bands on the agarose gel, so that we skipped this part of the project. | * Miniprep of NFAT responsive and NFkB responsive promoters and test digest at 37°C for one hour -> no right bands on the agarose gel, so that we skipped this part of the project. | ||
| - | == | + | == 9-22-2009 == |
* Tranformation of CYP1A1-muta. | * Tranformation of CYP1A1-muta. | ||
| - | * Analysis of the the following PCR products by agarose gel electrophoresis: RARE, LDLR, HMG CoA synthase and c-Jun promoter. | + | * Analysis of the the following PCR products by agarose gel electrophoresis: RARE, LDLR, HMG CoA synthase and c-Jun promoter (Fig. 4-5). |
| - | + | ||
* Digest of these PCR products by the restriction enzymes, NheI and EcoRI, at 37°C for one hour. | * Digest of these PCR products by the restriction enzymes, NheI and EcoRI, at 37°C for one hour. | ||
| - | [[image: | + | {| |
| + | |-valign="top" border="0" | ||
| + | |width="400px" style="background-color:#ede8e5; padding: 0 20px 0 0;"| | ||
| + | [[image:HD09_Addgene_teil1_1.png|center|600px|thumb|'''Figure 4: 1% agarose gel electrophoresis of the PCR products of LDLR, RARE and c-Jun promoter.''' The LDLR, RARE and c-Jun promoter PCR products were analysed by agarose gel electrophoresis. Two reaction mixtures were used; one with ''Taq'' polymerase and the other one with ''Phu'' polymerase. Additionally, a DNA ladder (100 - 10000 bp; Fermentas DNA Ladder Mix) is added into the gel for the validation of the DNA product length. The resulting c-Jun inserts should be 341 bp long, LDLR 363 bp and RARE 212 bp. All bands have the right length and could be used for cloning.]] | ||
| + | |width="450px"| | ||
| + | [[image:HD09_Addgene_teil2.2.png|center|600px|thumb|'''Figure 5: 1% agarose gel electrophoresis of the PCR products of HMG CoA synthase promoter.''' The HMG CoA synthase promoter PCR products were analysed by agarose gel electrophoresis. Two reaction mixtures were used; one with ''Taq'' polymerase and the other one with ''Phu'' polymerase. Additionally, a DNA ladder (100 - 10000 bp; Fermentas DNA Ladder Mix) is added into the gel for the validation of the DNA product length. The resulting HMG CoA synthase inserts should be 363 bp long. All bands have the right length and could be used for cloning.]] | ||
| + | |} | ||
| - | == | + | == 9-23-2009 == |
* Gel extraction of RARE, LDLR, HMG CoA synthase and c-Jun insert and combination of the two reaction mixtures (''Taq'' and ''Phu''). | * Gel extraction of RARE, LDLR, HMG CoA synthase and c-Jun insert and combination of the two reaction mixtures (''Taq'' and ''Phu''). | ||
* Ligation of these extracted inserts with the digested (NheI and EcoRI) plasmid p31 at 22°C for four hours. | * Ligation of these extracted inserts with the digested (NheI and EcoRI) plasmid p31 at 22°C for four hours. | ||
| Line 257: | Line 304: | ||
* Transformation of the RARE/p31, LDLR/p31, c-Jun/p31 and HMG CoA promoter/p31 construct. | * Transformation of the RARE/p31, LDLR/p31, c-Jun/p31 and HMG CoA promoter/p31 construct. | ||
| - | == | + | == 9-24-2009 == |
* Ligation of the HSP70 promoter with the digested (NheI and EcoRI) p31 plasmid at 22°C for three hours. | * Ligation of the HSP70 promoter with the digested (NheI and EcoRI) p31 plasmid at 22°C for three hours. | ||
* Transformation of the HSP70 promoter/p31 construct. | * Transformation of the HSP70 promoter/p31 construct. | ||
* Colonies of the RARE/p31, LDLR/p31, c-Jun/p31 and HMG CoA promoter/p31 construct are picked. | * Colonies of the RARE/p31, LDLR/p31, c-Jun/p31 and HMG CoA promoter/p31 construct are picked. | ||
| - | == | + | == 9-25-2009 == |
* Miniprep of RARE/p31, LDLR/p31, c-Jun/p31 and HMG CoA promoter/p31 construct. | * Miniprep of RARE/p31, LDLR/p31, c-Jun/p31 and HMG CoA promoter/p31 construct. | ||
* Colonies of the HSP70 promoter/p31 construct are picked. | * Colonies of the HSP70 promoter/p31 construct are picked. | ||
| - | == | + | == 9-26-2009 == |
* Miniprep of the HSP70 promoter/p31 construct. | * Miniprep of the HSP70 promoter/p31 construct. | ||
| - | * Test digest (NheI and EcoRI) of RARE, c-Jun, LDLR and HMG CoA construct at 37°C for one hour. | + | * Test digest (NheI and EcoRI) of RARE, c-Jun, LDLR and HMG CoA construct at 37°C for one hour (Fig. 6-10). |
| - | ''' | + | * Following constructs were send for sequencing: |
| - | [[image: | + | C-Jun #19, c-Jun #20, c-Jun #21, c-Jun #22, RARE #8, RARE #9, RARE #15, RARE #23, HMG CoA #24, HMG CoA #26, LDLR #3, LDLR #5, LDLR #11, LDLR #12, LDLR #19, LDLR #20, LDLR #21, LDLR #22, LDLR #24, LDLR #25, LDLR #26 and HMG CoA #35 . |
| + | |||
| + | [[image:HD09_Jun2.png|center|600px|thumb|'''Figure 6: 1% agarose gel electrophoresis of a test digest of the c-Jun/p31 plasmid constructs.'''The c-Jun/p31 plasmids were digested with EcoRI and PstI at 37°C for one hour. The numbers (1,3,4,5 etc.) describe the plasmids of different bacterial colonies. Additionally, a DNA ladder (100 - | ||
| + | 10000 bp; Fermentas DNA Ladder Mix) is added into the gel for the validation of the DNA product length. The resulting c-Jun insert should be 341 bp long. C-Jun #19, c-Jun #20, c-Jun #21 and c-Jun #22 seem to be successfully ligated plasmids, because there are small bands at ~ 300 bp. All lanes show a p31 plasmid band, which is 4919 bp long. But other c-Jun inserts could not be recognized and so all other lanes display definetly the wrong product (JeT, ~180 bp).]] | ||
| + | |||
| + | [[image:HD09_RARE.png|center|600px|thumb|'''Figure 7: 1% agarose gel electrophoresis of a test digest of the RARE/p31 plasmid constructs.'''The RARE/p31 plasmids were digested with EcoRI and PstI at 37°C for one hour. The numbers (1,3,4,5 etc.) describe the plasmids of different bacterial colonies. Additionally, a DNA ladder (100 - | ||
| + | 10000 bp; Fermentas DNA Ladder Mix) is added into the gel for the validation of the DNA product length. The resulting RARE insert should be 212 bp long. The RARE inserts are difficult to recognize, because the JeT insert of the original plasmid is ~ 180 bp. Therefore, some good constructs are chosen: RARE #8, RARE #9, RARE #15 and RARE #23.]] | ||
| + | |||
| + | [[image:HD09_HMGCoA.png|center|600px|thumb|'''Figure 8: 1% agarose gel electrophoresis of a test digest of the HMG CoA synthase/p31 plasmid constructs.'''The HMG CoA synthase/p31 plasmids were digested with EcoRI and PstI at 37°C for one hour. The numbers (1,3,4,5 etc.) describe the plasmids of different bacterial colonies. Additionally, a DNA ladder (100 - | ||
| + | 10000 bp; Fermentas DNA Ladder Mix) is added into the gel for the validation of the DNA product length. The resulting HMG CoA synthase insert should be 363 bp long. All lanes show a p31 plasmid band, which is 4919 bp long. But the HMG CoA inserts could not be recognized. All lanes display definetly the wrong product (JeT, ~180 bp).]] | ||
| + | |||
| + | [[image:HD09_LDL_HMG.png|center|600px|thumb|'''Figure 9: 1% agarose gel electrophoresis of a test digest of the LDLR/p31 and HMG CoA synthase/p31 plasmid constructs.'''The LDLR/p31 and HMG CoA synthase/p31 plasmids were digested with EcoRI and PstI at 37°C for one hour. The numbers (1,3,4,5 etc.) describe the plasmids of different bacterial colonies. Additionally, a DNA ladder (100 - | ||
| + | 10000 bp; Fermentas DNA Ladder Mix) is added into the gel for the validation of the DNA product length. The resulting LDLR insert should be 363 bp long and HMG CoA 363 bp. HMG CoA #24, HMG CoA #26, LDLR #3, LDLR #5, LDLR #11 and LDLR #12 seem to be successfully ligated plasmids, because there are small bands at ~ 350 bp. All lanes show a p31 plasmid band, which is 4919 bp long. But other HMG CoA and LDLR inserts could not be recognized. All other lanes display definetly the wrong product (JeT, ~180 bp).]] | ||
| + | |||
| + | [[image:HD09_HMG_LDL_2.png|center|600px|thumb|'''Figure 10: 1% agarose gel electrophoresis of a test digest of the LDLR/p31 and HMG CoA synthase/p31 plasmid constructs.'''The NFkB/p31 and NFAT/p31 plasmids were digested with EcoRI and PstI at 37°C for one hour. The numbers (1,3,4,5 etc.) describe the plasmids of different bacterial colonies. Additionally, a DNA ladder (100 - | ||
| + | 10000 bp; Fermentas DNA Ladder Mix) is added into the gel for the validation of the DNA product length. The resulting LDLR insert should be 363 bp long and HMG CoA 363 bp. LDLR #19, LDLR #20, LDLR #21, LDLR #22, LDLR #24, LDLR #25, LDLR #26 and HMG CoA #35 seem to be successfully ligated plasmids, because there are small bands at ~ 350 bp. All lanes show a p31 plasmid band, which is 4919 bp long. But other HMG CoA and LDLR inserts could not be recognized. All other lanes display definetly the wrong product (JeT, ~180 bp).]] | ||
| + | |||
| + | == 9-28-2009 == | ||
| + | * Right constructs of RARE, c-Jun, LDLR and HMG CoA are: c-Jun #19, c-Jun #20, c-Jun #21, c-Jun #22, RARE #8, RARE #23, LDLR #19, LDLR #20, LDLR #21 and HMG CoA #35. | ||
| + | * Test digest (NheI and EcoRI) of HSP70 at 37°C for one hour (Fig. 11). | ||
| + | * Following construct is send for sequencing: HSP70 #9. | ||
| - | |||
| - | [[image: | + | [[image:HD09_HSP70.png|center|600px|thumb|'''Figure 11: 1% agarose gel electrophoresis for a test digest of the HSP70/p31 plasmid construct.'''The HSP70/p31 plasmids were digested with EcoRI and PstI at 37°C for one hour. The numbers (1,3,4,5 etc.) describe the plasmids of different bacterial colonies. Additionally, a DNA ladder (100 - |
| + | 10000 bp; Fermentas DNA Ladder Mix) is added into the gel for the validation of the DNA product length. The resulting HSP70 insert should be 402 bp long. HSP70 #9 seem to be a successfully ligated plasmid, because there is a small bands at ~ 400 bp. All lanes show a p31 plasmid band, which is 4919 bp long. But other HSP70 inserts could not be recognized and so all other lanes display definetly the wrong product (JeT, ~180 bp).]] | ||
| - | + | == 9-29-2009 == | |
| + | * Right constructs of HSP70 is: HSP70 #9. | ||
| - | |||
| - | == | + | == 10-12-2009 == |
| - | * | + | * TECAN (plate reader) measurement of the natural LDL receptor promoter and the HMG CoA synthase promoter. These promoters, which are coupled to GFP, were cotransfected with a reference plasmid including JeT coupled to mCherry and were induced by Lipoprotein Deficient Serum (See: [[Team:Heidelberg/Notebook_promoters_cells| Cell Culture]]). |
| - | + | ||
| - | [[ | + | |
| - | | | + | [[image:HD09_LDL_HMG_CoA.png|center|400px|thumb|'''Figure 12: The HMG CoA synthase promoter and the LDL receptor promoter induced approximately 25-35% by Lipoprotein Deficient Serum.''' The LDL receptor promoter and HMG CoA synthase promoter is coupled to the fluorecent protein GFP. This GFP fluorescence was measured by TECAN once (à three replicates). Background fluorescence was substracted, and fluorescence levels are plotted relative to JeT. The standard deviations are indicated by the error bars.]] |
| + | '''Notebook information about the measurement of natural promoters is found in the Notebook [[Team:Heidelberg/Notebook_measure/NotebookFC| Measurement]] and [[Team:Heidelberg/Notebook_promoters_cells| Cell Culture]].''' | ||
|} | |} | ||
Latest revision as of 11:43, 20 October 2009
Natural PromotersContents
8-11-2009
8-18-2009
8-19-2009
8-20-2009
8-21-2009
8-24-2009
8-25-2009
8-26-2009
 Figure 1: 1% agarose gel electrophoresis of the PCR products of APOAIV and CYP1A1 promoter.The APOAIV and CYP1A1 promoter PCR products were analysed by agarose gel electrophoresis. Two reaction mixtures were used; one with Taq polymerase and the other one with Phu polymerase and with and without DMSO adding. Besides, the reaction mixtures of Phu have a varying concentration of MGCl2 (1-4 µl in a 50 µl reaction mixture). Additionally, a DNA ladder (100 - 10000 bp; Fermentas DNA Ladder Mix) is added into the gel for the validation of the DNA product length. The resulting CYP1A1 inserts should be 1229 bp long and APOAIV ~4200 bp. The "CYP1A1 Taq" and "CYP1A1 Taq + DMSO" are the only products, which have the right length. All the other lanes show an unspecific PCR. 8-27-2009
8-28-2009
8-29-2009
8-31-2009
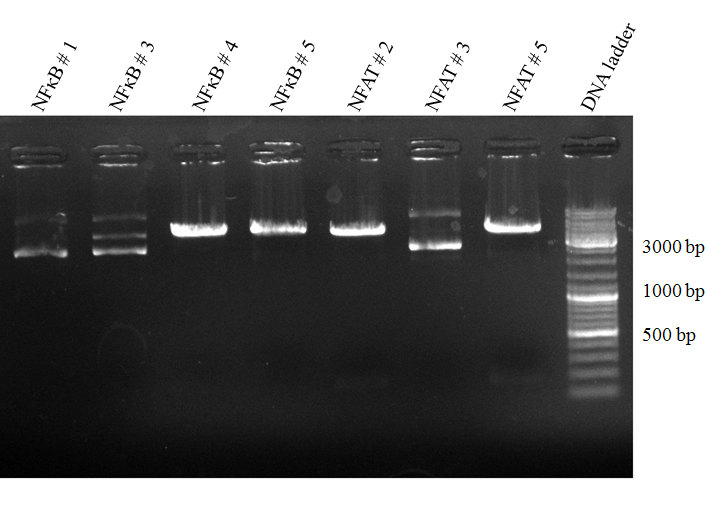 Figure 2: 1% agarose gel electrophoresis for a test digest of the NFkB/p31 plasmid and the NFAT/p31 plasmid. The NFkB/p31 and NFAT/p31 plasmids were digested with EcoRI and PstI at 37°C for one hour. The numbers (1,3,4,5 etc.) describe the plasmids of different bacterial colonies. Additionally, a DNA ladder (100 - 10000 bp; Fermentas DNA Ladder Mix) is added into the gel for the validation of the DNA product length. The resulting NFAT insert should be 134 bp long and NFkB 179 bp. NFAT #2 and NFAT #5 seem to be successfully ligated plasmids, because there are small bands at ~ 150 bp. The lanes of NFkB #4 and NFkB #5 show a p31 plasmid band, which is 4919 bp long. But the NFkB inserts could not be recognized. Nevertheless, we will try to sequence them. All other lanes display definetly the wrong products. 9-01-2009
9-02-2009
9-03-2009
9-04-2009
9-07-2009
9-08-2009
9-09-2009
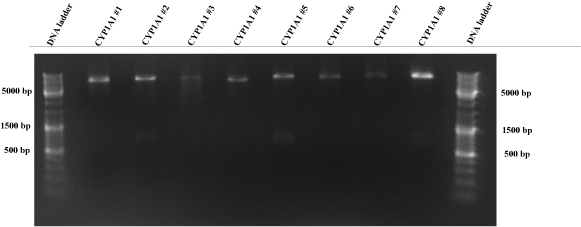 Figure 3: 1% agarose gel electrophoresis for a test digest of the CYP1A1/p31 plasmid. The CYP1A1/p31 plasmids were digested with EcoRI and PstI at 37°C for one hour. The numbers (1,3,4,5 etc.) describe the plasmids of different bacterial colonies. Additionally, a DNA ladder (75 -20000 bp); Fermentas DNA 1kb Plus) is added into the gel for the validation of the DNA product length. The resulting CYP1A1 insert should be 1229 bp long. CYP1A1 #2, CYP1A1 #5 and CYP1A1 #8 seem to be successfully ligated plasmids, because there are small bands at ~ 1200 bp. All lanes show a p31 plasmid band, which is 4919 bp long. But other CYP1A1 inserts could not be recognized. All other lanes display definetly the wrong product (JeT, ~180 bp). 9-10-09
9-11-09
9-14-2009
9-15-2009
9-16-2009
9-17-2009
9-18-2009
9-20-2009
9-21-2009
9-22-2009
9-23-2009
9-24-2009
9-25-2009
9-26-2009
C-Jun #19, c-Jun #20, c-Jun #21, c-Jun #22, RARE #8, RARE #9, RARE #15, RARE #23, HMG CoA #24, HMG CoA #26, LDLR #3, LDLR #5, LDLR #11, LDLR #12, LDLR #19, LDLR #20, LDLR #21, LDLR #22, LDLR #24, LDLR #25, LDLR #26 and HMG CoA #35 . 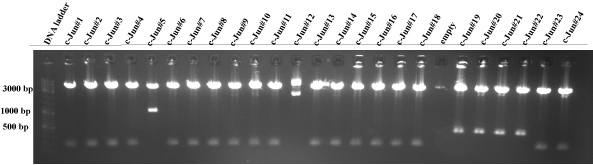 Figure 6: 1% agarose gel electrophoresis of a test digest of the c-Jun/p31 plasmid constructs.The c-Jun/p31 plasmids were digested with EcoRI and PstI at 37°C for one hour. The numbers (1,3,4,5 etc.) describe the plasmids of different bacterial colonies. Additionally, a DNA ladder (100 - 10000 bp; Fermentas DNA Ladder Mix) is added into the gel for the validation of the DNA product length. The resulting c-Jun insert should be 341 bp long. C-Jun #19, c-Jun #20, c-Jun #21 and c-Jun #22 seem to be successfully ligated plasmids, because there are small bands at ~ 300 bp. All lanes show a p31 plasmid band, which is 4919 bp long. But other c-Jun inserts could not be recognized and so all other lanes display definetly the wrong product (JeT, ~180 bp). 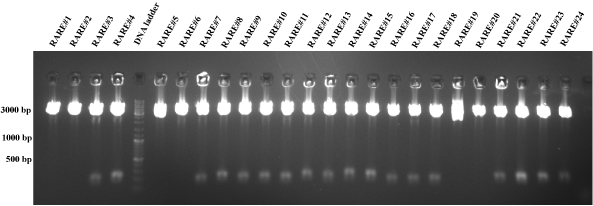 Figure 7: 1% agarose gel electrophoresis of a test digest of the RARE/p31 plasmid constructs.The RARE/p31 plasmids were digested with EcoRI and PstI at 37°C for one hour. The numbers (1,3,4,5 etc.) describe the plasmids of different bacterial colonies. Additionally, a DNA ladder (100 - 10000 bp; Fermentas DNA Ladder Mix) is added into the gel for the validation of the DNA product length. The resulting RARE insert should be 212 bp long. The RARE inserts are difficult to recognize, because the JeT insert of the original plasmid is ~ 180 bp. Therefore, some good constructs are chosen: RARE #8, RARE #9, RARE #15 and RARE #23.  Figure 8: 1% agarose gel electrophoresis of a test digest of the HMG CoA synthase/p31 plasmid constructs.The HMG CoA synthase/p31 plasmids were digested with EcoRI and PstI at 37°C for one hour. The numbers (1,3,4,5 etc.) describe the plasmids of different bacterial colonies. Additionally, a DNA ladder (100 - 10000 bp; Fermentas DNA Ladder Mix) is added into the gel for the validation of the DNA product length. The resulting HMG CoA synthase insert should be 363 bp long. All lanes show a p31 plasmid band, which is 4919 bp long. But the HMG CoA inserts could not be recognized. All lanes display definetly the wrong product (JeT, ~180 bp). 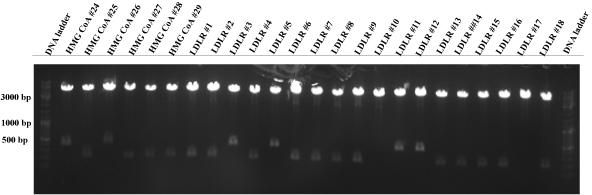 Figure 9: 1% agarose gel electrophoresis of a test digest of the LDLR/p31 and HMG CoA synthase/p31 plasmid constructs.The LDLR/p31 and HMG CoA synthase/p31 plasmids were digested with EcoRI and PstI at 37°C for one hour. The numbers (1,3,4,5 etc.) describe the plasmids of different bacterial colonies. Additionally, a DNA ladder (100 - 10000 bp; Fermentas DNA Ladder Mix) is added into the gel for the validation of the DNA product length. The resulting LDLR insert should be 363 bp long and HMG CoA 363 bp. HMG CoA #24, HMG CoA #26, LDLR #3, LDLR #5, LDLR #11 and LDLR #12 seem to be successfully ligated plasmids, because there are small bands at ~ 350 bp. All lanes show a p31 plasmid band, which is 4919 bp long. But other HMG CoA and LDLR inserts could not be recognized. All other lanes display definetly the wrong product (JeT, ~180 bp).  Figure 10: 1% agarose gel electrophoresis of a test digest of the LDLR/p31 and HMG CoA synthase/p31 plasmid constructs.The NFkB/p31 and NFAT/p31 plasmids were digested with EcoRI and PstI at 37°C for one hour. The numbers (1,3,4,5 etc.) describe the plasmids of different bacterial colonies. Additionally, a DNA ladder (100 - 10000 bp; Fermentas DNA Ladder Mix) is added into the gel for the validation of the DNA product length. The resulting LDLR insert should be 363 bp long and HMG CoA 363 bp. LDLR #19, LDLR #20, LDLR #21, LDLR #22, LDLR #24, LDLR #25, LDLR #26 and HMG CoA #35 seem to be successfully ligated plasmids, because there are small bands at ~ 350 bp. All lanes show a p31 plasmid band, which is 4919 bp long. But other HMG CoA and LDLR inserts could not be recognized. All other lanes display definetly the wrong product (JeT, ~180 bp). 9-28-2009
 Figure 11: 1% agarose gel electrophoresis for a test digest of the HSP70/p31 plasmid construct.The HSP70/p31 plasmids were digested with EcoRI and PstI at 37°C for one hour. The numbers (1,3,4,5 etc.) describe the plasmids of different bacterial colonies. Additionally, a DNA ladder (100 - 10000 bp; Fermentas DNA Ladder Mix) is added into the gel for the validation of the DNA product length. The resulting HSP70 insert should be 402 bp long. HSP70 #9 seem to be a successfully ligated plasmid, because there is a small bands at ~ 400 bp. All lanes show a p31 plasmid band, which is 4919 bp long. But other HSP70 inserts could not be recognized and so all other lanes display definetly the wrong product (JeT, ~180 bp). 9-29-2009
10-12-2009
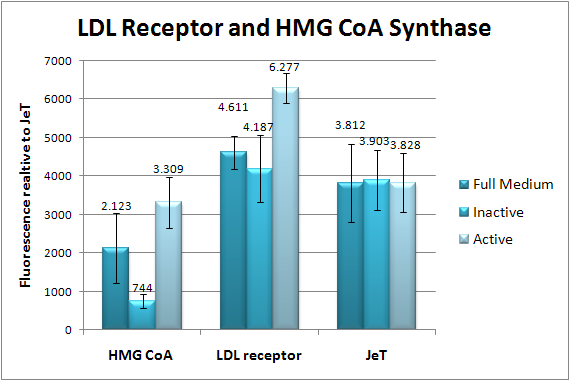 Figure 12: The HMG CoA synthase promoter and the LDL receptor promoter induced approximately 25-35% by Lipoprotein Deficient Serum. The LDL receptor promoter and HMG CoA synthase promoter is coupled to the fluorecent protein GFP. This GFP fluorescence was measured by TECAN once (à three replicates). Background fluorescence was substracted, and fluorescence levels are plotted relative to JeT. The standard deviations are indicated by the error bars. Notebook information about the measurement of natural promoters is found in the Notebook Measurement and Cell Culture. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"

