EPF-Lausanne/18 October 2009
From 2009.igem.org
(→People in the lab) |
(→Wet Lab) |
||
| (8 intermediate revisions not shown) | |||
| Line 26: | Line 26: | ||
==Wet Lab== | ==Wet Lab== | ||
| + | '''Broth culture''' | ||
| + | We did a culture of every plate from yesterday (only the 4x diluted, because otherwise it was too concentrated). RO1 transf., RO2 transf., D transf. RO2.4 + ILE 427 PHE #10, D transf. RO1.1 +ILE 427 PHE #10, RO2.4 + BB1, RO1.1 + BB1. Finally this culture has not been used. | ||
| + | |||
| + | The following clones have been put in culture for the afternoon characterization : 2ml LB + antibiotic + 2 mM IPTG, tips from plates : | ||
| + | |||
| + | RO1.1 + BB1 #3 (from the plate), RO2.4 + BB1 #3 (the original, from overnight culture, 10ul inoculation), RO2.4 + BB1 #2 (plate), RO1.1 + BB1' #1,2,3 and RO2.4 + BB1' #1,3,6 with mutated LovTAP gene : ILE 427 PHE. | ||
| + | |||
| + | We also did a '''glycerol stock''' of ILE 427 PHE #1 and #10. | ||
| + | |||
| + | We then did a '''colony PCR''', using the protocol with Taq platinium. We did the following clones : RO1.1 + ILE 427 PHE #10 dil. 4x n° 1,2,3,4,5,6; D transf RO2.4 + ILE 427 PHE #10 dil. 4x n°1,2,3,4,5,6; RO1.1 + BB1 dil. 4x 1,2,3; RO2.4 + BB1 dil. 4x 1,2,3,4. We then did a '''gel of the colony PCR'''. The gel is curved, so not too good. Morever it has a strange colour... we will do a new gel tomorrow ! From what we can see, RO2.4 is missing, RO1.1 seems okay (lengths are right) but not for all the clones (RO1.1 + BB1' #3,4,5,6 okay, RO1.1 + BB1 #13,14,15 okay). | ||
| + | |||
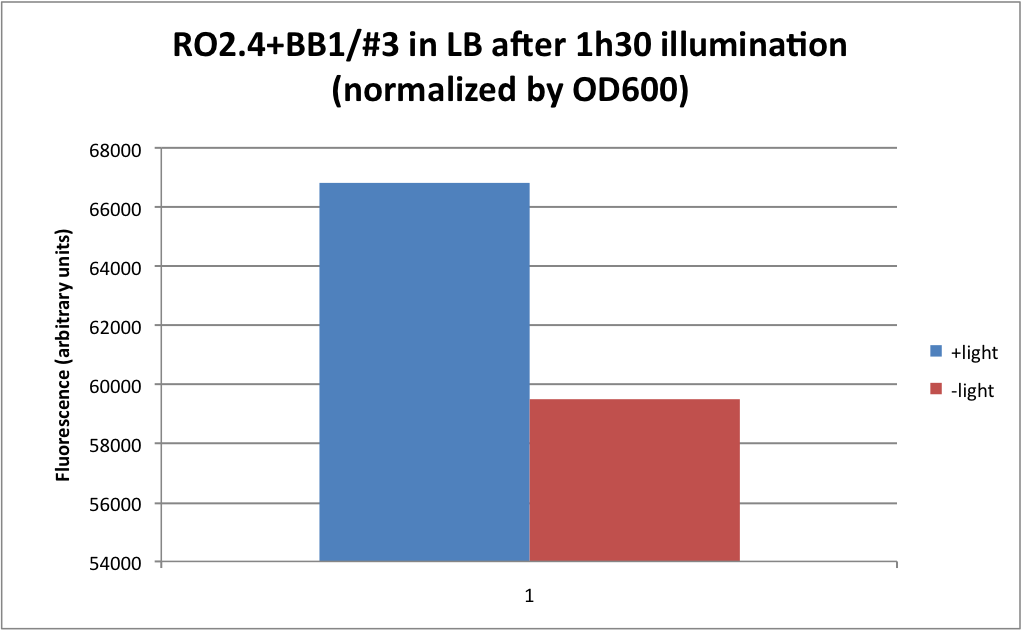
| + | We then put into '''culture''' some other clones : the "tips induced" cultures were very low... it is quite normal as we were not able to take a lot out of the plates. After about 3h30, all tubes were clear (except RO2.4 + BB1 "original"). We switched the light on for 1h30 (for +light clones). We did the plate experiment even though we couldn't see any turbidity in the tubes (except "origninal" RO2.4+BB1) | ||
| + | |||
| + | |||
| + | |||
| + | '''Characterisation experiment''' : | ||
| + | |||
| + | We did two experiments : | ||
| + | |||
| + | 1. Static with about 1h30 light induction and negative control in the dark. | ||
| + | |||
| + | 2. Experiment to assess the stability of mutants. They were grown in the dark and then induced for 45 s by light. Try the difference in fluorescence evolution between BB1 and BB1'. | ||
| + | |||
| + | Note : both plates were done at the same time. Loaded in the dark under red LEDs. | ||
| + | |||
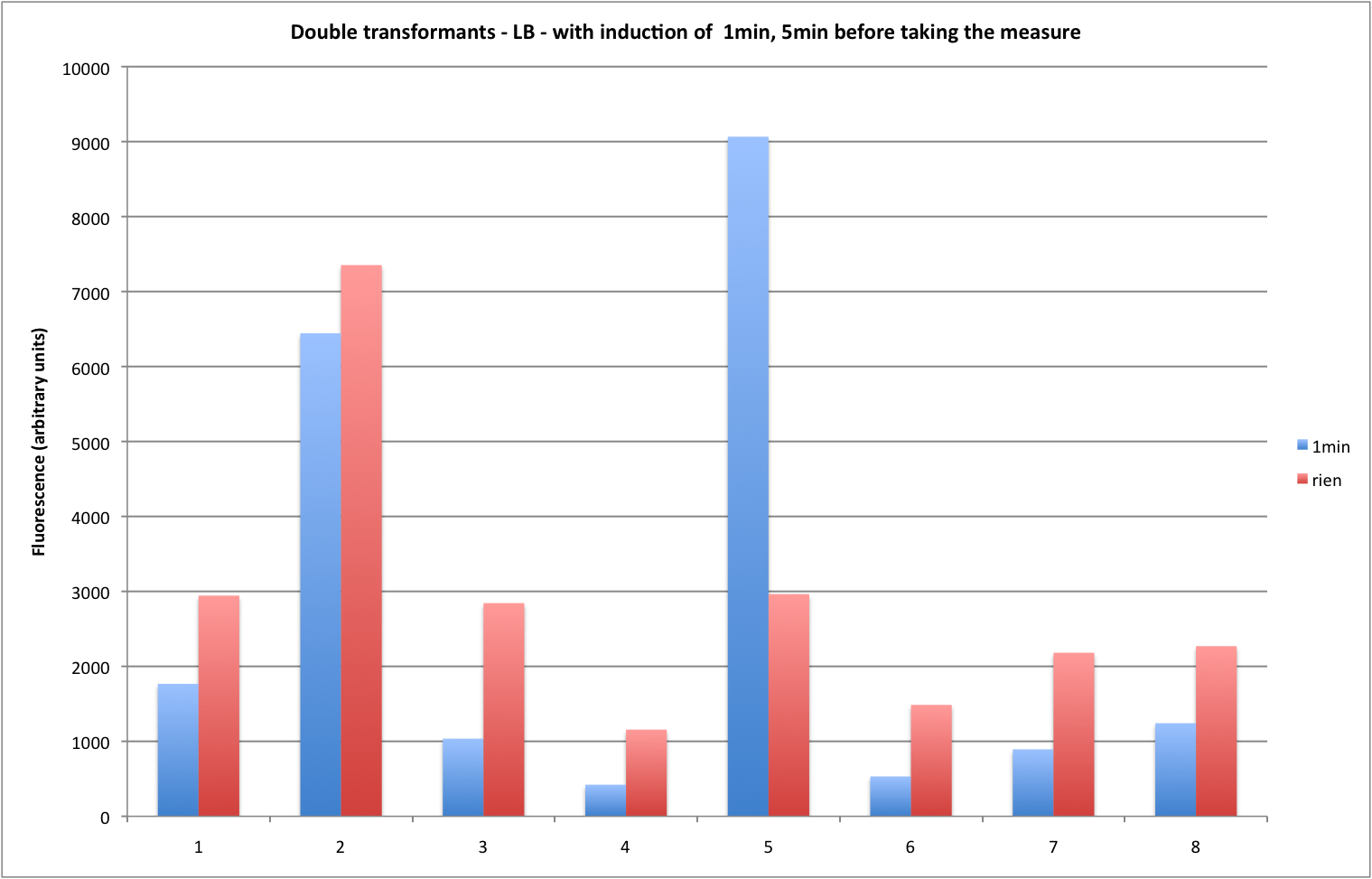
| + | For the stability assess : 1 minutes of light was shined on the plate just before taking them to the plate reader. | ||
| + | |||
| + | '''Results''' | ||
| + | |||
| + | |||
| + | [[Image:dt_1min.png|center|thumb|upright=4|Double transformants]] | ||
| + | |||
| + | [[Image:dt_static.png|center|thumb|upright=4|Double transformants : static experiment]] | ||
| + | |||
| + | [[Image:RO2.4+BB1_1min.png|center|thumb|upright=4|RO2.4 + BB1]] | ||
| + | |||
| + | [[Image:181009_.png|center|thumb|upright=4|181009]] | ||
| + | |||
| + | |||
| + | Conclusion of these experiments : | ||
| + | |||
| + | Columns : | ||
| + | |||
| + | 1=RO1.1+BB1/3-->okay if we look at the gel | ||
| + | |||
| + | 2=RO1.1+BB1(mutated)/1-->not okay | ||
| + | |||
| + | 3=RO1.1+BB1(mutated)/2-->not okay | ||
| + | |||
| + | 4=RO1.1+BB1(mutated)/3-->okay | ||
| + | |||
| + | 5=RO2.4+BB1/2-->not okay | ||
| + | |||
| + | 6=RO2.4+BB1(muté)/1-->not okay | ||
| + | |||
| + | 7=RO2.4+BB1(muté)/3-->not okay | ||
| + | |||
| + | 8=RO2.4+BB1(muté)/6-->maybe okay ? | ||
| + | |||
| + | The cultures weren't even turbid when we did the measures. This could explain why we got such strange measures and not what we expected. No real conclusion can be taken from these results and further experiment should be done in order to assess if the BB1' isn't really functional. Only RO2.4 + BB1 "original" was okay. So at least we know that this experiment is reproducible and that our BB1 (and thus LovTAP protein) is indeed functional. | ||
==People in the lab== | ==People in the lab== | ||
Latest revision as of 12:43, 21 October 2009
Contents |
Wet Lab
Broth culture
We did a culture of every plate from yesterday (only the 4x diluted, because otherwise it was too concentrated). RO1 transf., RO2 transf., D transf. RO2.4 + ILE 427 PHE #10, D transf. RO1.1 +ILE 427 PHE #10, RO2.4 + BB1, RO1.1 + BB1. Finally this culture has not been used.
The following clones have been put in culture for the afternoon characterization : 2ml LB + antibiotic + 2 mM IPTG, tips from plates :
RO1.1 + BB1 #3 (from the plate), RO2.4 + BB1 #3 (the original, from overnight culture, 10ul inoculation), RO2.4 + BB1 #2 (plate), RO1.1 + BB1' #1,2,3 and RO2.4 + BB1' #1,3,6 with mutated LovTAP gene : ILE 427 PHE.
We also did a glycerol stock of ILE 427 PHE #1 and #10.
We then did a colony PCR, using the protocol with Taq platinium. We did the following clones : RO1.1 + ILE 427 PHE #10 dil. 4x n° 1,2,3,4,5,6; D transf RO2.4 + ILE 427 PHE #10 dil. 4x n°1,2,3,4,5,6; RO1.1 + BB1 dil. 4x 1,2,3; RO2.4 + BB1 dil. 4x 1,2,3,4. We then did a gel of the colony PCR. The gel is curved, so not too good. Morever it has a strange colour... we will do a new gel tomorrow ! From what we can see, RO2.4 is missing, RO1.1 seems okay (lengths are right) but not for all the clones (RO1.1 + BB1' #3,4,5,6 okay, RO1.1 + BB1 #13,14,15 okay).
We then put into culture some other clones : the "tips induced" cultures were very low... it is quite normal as we were not able to take a lot out of the plates. After about 3h30, all tubes were clear (except RO2.4 + BB1 "original"). We switched the light on for 1h30 (for +light clones). We did the plate experiment even though we couldn't see any turbidity in the tubes (except "origninal" RO2.4+BB1)
Characterisation experiment :
We did two experiments :
1. Static with about 1h30 light induction and negative control in the dark.
2. Experiment to assess the stability of mutants. They were grown in the dark and then induced for 45 s by light. Try the difference in fluorescence evolution between BB1 and BB1'.
Note : both plates were done at the same time. Loaded in the dark under red LEDs.
For the stability assess : 1 minutes of light was shined on the plate just before taking them to the plate reader.
Results
Conclusion of these experiments :
Columns :
1=RO1.1+BB1/3-->okay if we look at the gel
2=RO1.1+BB1(mutated)/1-->not okay
3=RO1.1+BB1(mutated)/2-->not okay
4=RO1.1+BB1(mutated)/3-->okay
5=RO2.4+BB1/2-->not okay
6=RO2.4+BB1(muté)/1-->not okay
7=RO2.4+BB1(muté)/3-->not okay
8=RO2.4+BB1(muté)/6-->maybe okay ?
The cultures weren't even turbid when we did the measures. This could explain why we got such strange measures and not what we expected. No real conclusion can be taken from these results and further experiment should be done in order to assess if the BB1' isn't really functional. Only RO2.4 + BB1 "original" was okay. So at least we know that this experiment is reproducible and that our BB1 (and thus LovTAP protein) is indeed functional.
People in the lab
Heidi, Basile, Caroline


 "
"