Team:UNICAMP-Brazil/Coliguard/Recognition
From 2009.igem.org
| Line 1: | Line 1: | ||
{{:Team:UNICAMP-Brazil/inc_topo}} | {{:Team:UNICAMP-Brazil/inc_topo}} | ||
| - | + | =The Coliguard - Recognition= | |
| - | + | ==Introduction== | |
We aim to create bacteria that can protect themselves against contaminants. So, our bacteria need to be able to kill other organisms which may appear in the culture medium. | We aim to create bacteria that can protect themselves against contaminants. So, our bacteria need to be able to kill other organisms which may appear in the culture medium. | ||
| Line 27: | Line 27: | ||
• But, what if the contaminants do not produce AI-2? | • But, what if the contaminants do not produce AI-2? | ||
| + | |||
In these situations, our E.coli will use another recognition mechanism, based in conjugation. In conjugation, a donor bacterium only conjugates with organisms that don’t have the same conjugative plasmid. Bacteria that already have the plasmid display membrane proteins that prevent the conjugation with other bacteria that have the same conjugative plasmid. (reference) Therefore, if our E.coli has a conjugative plasmid, they will only conjugate with different organisms, which will be the contaminants. | In these situations, our E.coli will use another recognition mechanism, based in conjugation. In conjugation, a donor bacterium only conjugates with organisms that don’t have the same conjugative plasmid. Bacteria that already have the plasmid display membrane proteins that prevent the conjugation with other bacteria that have the same conjugative plasmid. (reference) Therefore, if our E.coli has a conjugative plasmid, they will only conjugate with different organisms, which will be the contaminants. | ||
| Line 50: | Line 51: | ||
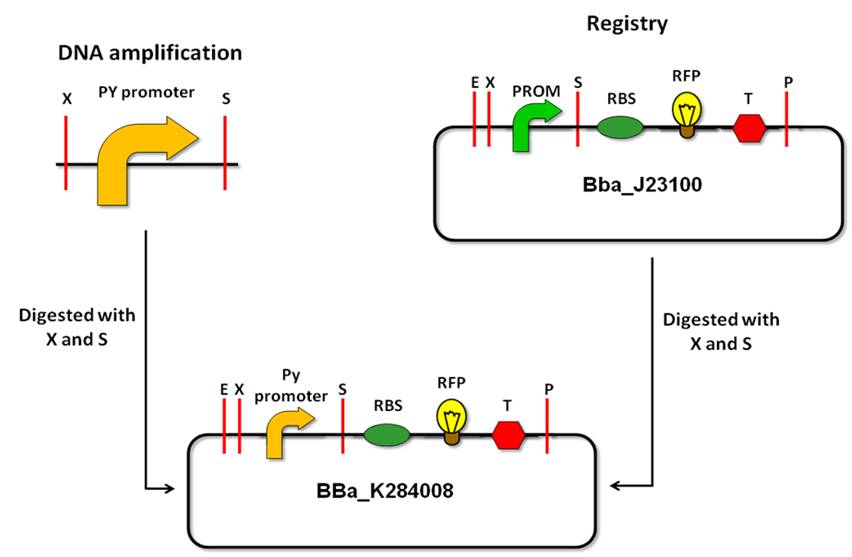
| - | Py promoter isolation | + | ==Py promoter isolation== |
| - | Strategy | + | '''Strategy''' |
The strategy consists of PCR amplification of the Py promoter region by using primers that contain Biobrick prefix and Biobrick suffix, thus allowing us to clone the amplicon as Biobrick parts. Before designing primers, we verified that none of the restriction sites of the prefix or sufixe are contained in the amplified sequence. | The strategy consists of PCR amplification of the Py promoter region by using primers that contain Biobrick prefix and Biobrick suffix, thus allowing us to clone the amplicon as Biobrick parts. Before designing primers, we verified that none of the restriction sites of the prefix or sufixe are contained in the amplified sequence. | ||
| Line 63: | Line 64: | ||
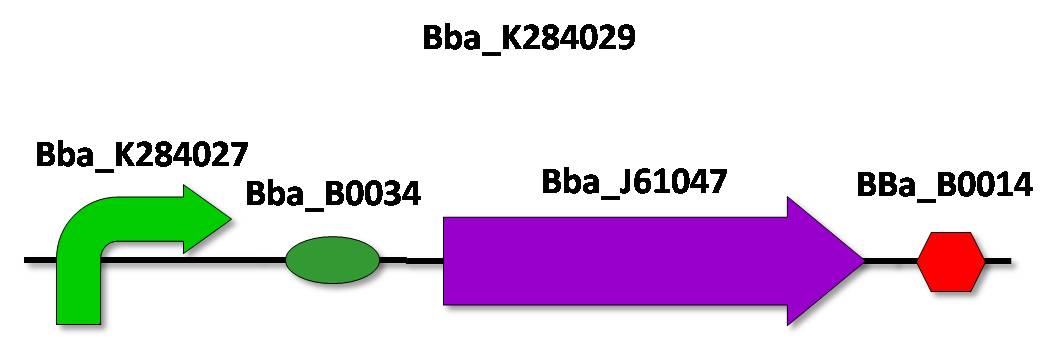
| - | + | ==Py promoter cloning and characterization== | |
| - | Strategy | + | '''Strategy''' |
After isolation of the Py promoter, it must be cloned in the BioBrick vector according to the standard assembly strategy. Afterwards, to characterize the promoter activity, we will make a device, with a reporter gene under the control of the Py promoter. We decided to use a fluorescent protein as a reporter gene for this characterization, such as the GFP (Green Fluorescent Protein). This reporter system will allow us to use the fluorescence signal in order to quantify promoter activity in a spectrofluorometer. (reference) | After isolation of the Py promoter, it must be cloned in the BioBrick vector according to the standard assembly strategy. Afterwards, to characterize the promoter activity, we will make a device, with a reporter gene under the control of the Py promoter. We decided to use a fluorescent protein as a reporter gene for this characterization, such as the GFP (Green Fluorescent Protein). This reporter system will allow us to use the fluorescence signal in order to quantify promoter activity in a spectrofluorometer. (reference) | ||
| Line 84: | Line 85: | ||
In the characterization experiment, we will use a donor bacterium that carries an F plasmid with the ampicillin resistance gene. Thus, we can’t select transformed bacterial colonies with our device in a medium supplemented with ampicillin. Therefore, we cloned the BBa_K284008 part into the pSB1AK3 vector, which carries the ampicillin and kanamycin resistance genes. Then, we could select the colonies in a medium supplemented with kanamycin. | In the characterization experiment, we will use a donor bacterium that carries an F plasmid with the ampicillin resistance gene. Thus, we can’t select transformed bacterial colonies with our device in a medium supplemented with ampicillin. Therefore, we cloned the BBa_K284008 part into the pSB1AK3 vector, which carries the ampicillin and kanamycin resistance genes. Then, we could select the colonies in a medium supplemented with kanamycin. | ||
| - | + | ==Engineering the F plasmid – removing parts and adding new replication origins== | |
| - | Strategy | + | '''Strategy''' |
First of all, we will reduce the size of the F plasmid by removing uninteresting parts and the insertion sequences, which are the portions of the F plasmid DNA that mediate the integration into the bacterial chromosome (generating Hfr strains). As Willetts and Johnson reported (reference), we will remove a part from the F plasmid by single digestion with HindIII followed by recircularization. This process creates pED100, a conjugative F plasmid derivative without insertion sequences. Through this simple procedure we will reduce considerably the possibility of plasmid integration into the chromosome. | First of all, we will reduce the size of the F plasmid by removing uninteresting parts and the insertion sequences, which are the portions of the F plasmid DNA that mediate the integration into the bacterial chromosome (generating Hfr strains). As Willetts and Johnson reported (reference), we will remove a part from the F plasmid by single digestion with HindIII followed by recircularization. This process creates pED100, a conjugative F plasmid derivative without insertion sequences. Through this simple procedure we will reduce considerably the possibility of plasmid integration into the chromosome. | ||
| Line 95: | Line 96: | ||
We managed to remove the insertions sequences and the uninteresting parts from the F plasmid. At this way, our plasmid is now easier to copy and to work with. | We managed to remove the insertions sequences and the uninteresting parts from the F plasmid. At this way, our plasmid is now easier to copy and to work with. | ||
| - | The recognition by AI-2 | + | ==The recognition by AI-2== |
| - | Strategy | + | |
| + | '''Strategy''' | ||
| + | |||
A well-known mechanism of cell-cell communication is called quorum sensing. In that process, bacteria communicate via secreted signaling molecules called autoinducers, which contribute to the regulation of the expression of particular genes. This is found in both gram-negative and gram-positive bacteria (1; 2). The most widespread quorum-sensing system is the luxS system (3). This inducer has been found in a large range of gram-negative and gram-positive bacteria, including some pathogens (4). Because this autoinducer and its biosynthetic pathway are the same among all bacterial species that possess luxS, it has been proposed that this system could be used in interspecies communication (5). AI-2 is produced from S-adenosyl-L-methionine (SAM) in three enzymatic steps. First, SAM acts as a methyl donor in SAM-dependent methyltransferase reactions, which lead to the formation of S-adenosyl-L-homocysteine (SAH). The next step is catalyzed by a nucleosidase (Pfs) encoded by the pfs gene that cleaves the glycosidic bond in both 50-methylthioadenosine (MTA) and (SAH). When SAH serves as a substrate, adenine and S-ribosyl-L-homocysteine (SRH) are formed. LuxS finally converts SRH into homocysteine and 4,5-dihydroxy-2,3-pentanedione (DPD). DPD is very unstableand, reacts with water and cyclizes forming autoinducer-2 (AI-2) (6). | A well-known mechanism of cell-cell communication is called quorum sensing. In that process, bacteria communicate via secreted signaling molecules called autoinducers, which contribute to the regulation of the expression of particular genes. This is found in both gram-negative and gram-positive bacteria (1; 2). The most widespread quorum-sensing system is the luxS system (3). This inducer has been found in a large range of gram-negative and gram-positive bacteria, including some pathogens (4). Because this autoinducer and its biosynthetic pathway are the same among all bacterial species that possess luxS, it has been proposed that this system could be used in interspecies communication (5). AI-2 is produced from S-adenosyl-L-methionine (SAM) in three enzymatic steps. First, SAM acts as a methyl donor in SAM-dependent methyltransferase reactions, which lead to the formation of S-adenosyl-L-homocysteine (SAH). The next step is catalyzed by a nucleosidase (Pfs) encoded by the pfs gene that cleaves the glycosidic bond in both 50-methylthioadenosine (MTA) and (SAH). When SAH serves as a substrate, adenine and S-ribosyl-L-homocysteine (SRH) are formed. LuxS finally converts SRH into homocysteine and 4,5-dihydroxy-2,3-pentanedione (DPD). DPD is very unstableand, reacts with water and cyclizes forming autoinducer-2 (AI-2) (6). | ||
Revision as of 19:06, 21 October 2009
| ||||||||||||||||||||||||||||||||||
 "
"