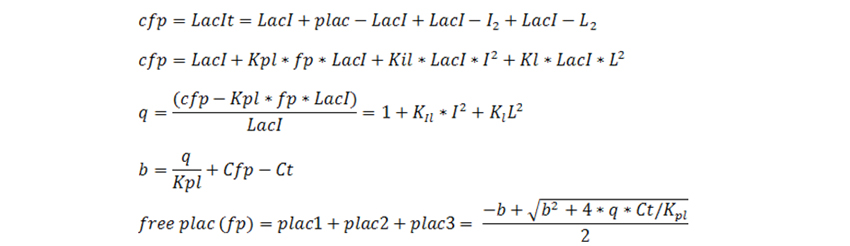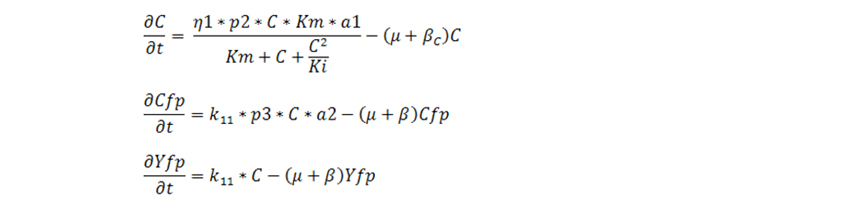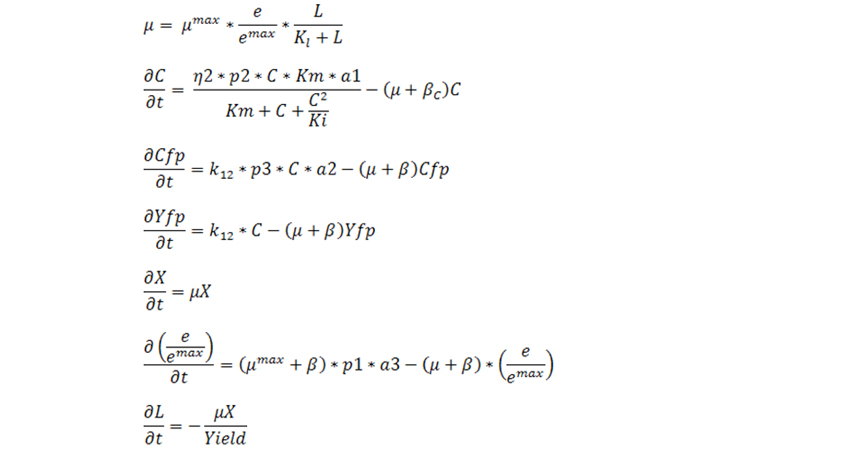Team:IIT Bombay India/DDM
From 2009.igem.org
Pranayiitb (Talk | contribs) |
Pranayiitb (Talk | contribs) |
||
| Line 22: | Line 22: | ||
|} | |} | ||
| - | {| background-color:#ffffff;" cellpadding=" | + | {| background-color:#ffffff;" cellpadding="2" cellspacing="1" border="0" bordercolor="#ffffff" width="90%" align="center" |
!align="left"| | !align="left"| | ||
| | | | ||
Revision as of 20:10, 21 October 2009
| Home | The Team | The Project | Analysis | Modeling | Notebook | Safety |
|---|
Detailed Deterministic Model |
|
Detailed Molecular Model Objective Here we wish to show how the dynamics of the cellular material (proteins and plasmids) changes with time and IPTG and also how the specific growth rate of the four constructs on lactose is controlled and maximized by use of multiple feedbacks. In this model quantification by simulation was done and later results were verified by experimental data.
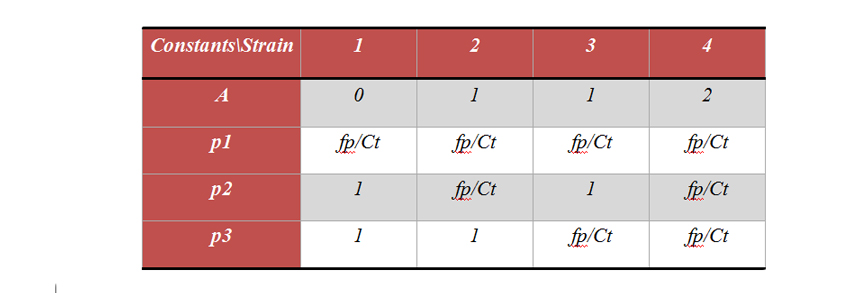
In our system we have the key components being plasmid copy number, fusion protein, yfp, lactose, IPTG and growth associated enzyme β-galactosidase. The E. coli genome inherently consists of β gal gene which has plac promoter. LacI interacts with lactose and IPTG and also with plac promoter. Assuming these 3 equilibrium reactions, we can now write differential equations for the components relating their concentrations with time. The total amount of plac promoter present in any strain could be given by the equation: Where ‘a’ is an integer which depends on the strain for which differential equation has been used to describe.(Total plac promoter, is the sum of concentration of free plac(fp) promoter and plac-LacI complex.
LacI total equals cfp (because they are a fusion protein). LacI refers to unbounded free lacI in the medium.
Note: here plac1, plac2, plac3 are the free plac associated with β-gal production, plasmid number and cfp-LacI protein. The differential equations are solved for two different conditions. Equations were first solved for 24 hours on other medium with different IPTG and no lactose. After 24 hours the equations were solved for the same value of IPTG but on different values of lactose. Equations for growth on no Lactose:
Equations for growth on Lactose:
The simulated results for the 4 strains are discussed below. The results are characterized in three parts; growth on non lactose media, subsequent growth on lactose and then we show how the multiple feedback helps in increasing growth rate of cells with reduced burden for production of proteins
|
 "
"