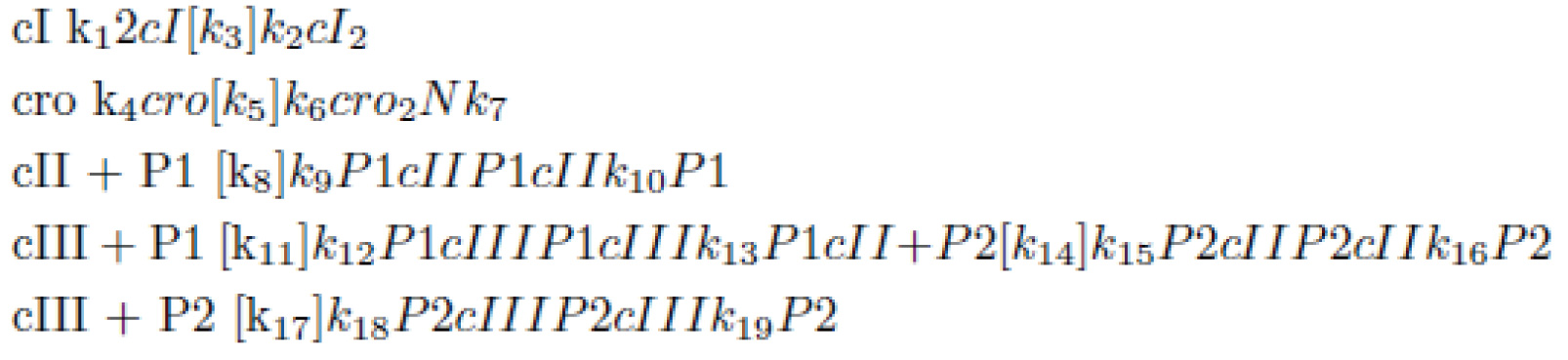Team:Todai-Tokyo/Modeling
From 2009.igem.org
M.Salvador (Talk | contribs) (→Modeling and the Result) |
|||
| (37 intermediate revisions not shown) | |||
| Line 55: | Line 55: | ||
== '''Overview''' == | == '''Overview''' == | ||
| - | As we described in the project overview of the bioclock project, the actual clock consists of two separable parts: a bio-oscillator and a lambda phage switch. In | + | As we described in the project overview of the bioclock project, the actual clock consists of two separable parts: a bio-oscillator and a lambda phage switch. In this section we will be modeling each of the parts and have come up with an integrated model, resembling the ultimate goal of the project. By making comparisons between the results of our model and the results of the original article which we based our simulation upon, we examined how realistic the model is, and tried to gain a better understanding of how the oscillation process and, eventually, how the integrated model works. |
| - | + | ||
== '''Bio-Oscillator''' == | == '''Bio-Oscillator''' == | ||
=== Construction of the Model === | === Construction of the Model === | ||
| - | In the natural world, there are quite a few | + | In the natural world, there are quite a few systems that are known to act as an oscillator. The project started out by examining the behavior of Cyanobacteria. We learnt that the mutual activation and repression between several compounds resulted in the oscillation pattern of protein expression. We finally settled on creating a system utilizing lacI and araC as the mutual-interaction compound to construct a oscillator. |
| - | Although it is possible to trace the reactions in detail all the way from the transcription of mRNA and translation of the proteins | + | Although it is possible to trace the reactions in detail all the way from the transcription of mRNA and translation of the proteins with the precise reaction rates, it will be redundant to do so. In order to maintain clarity, we simplify this process to focus only on the rate of translation. One of the defining characteristics is the dual role of the promotor. It could be bound to a maximum of two lacI proteins and one araC protien. When araC is bound to it, it activates the translation of both araC and lacI, increasing lacI concentration while enhancing the possibility of lacI binding to the promotor as well. When the promotor is bound to lacI, it could transmute in to a looped form, to which araC cannot bind. We will note the state of promotor as [[Image:suusiki_1.jpg|50px]], where i represents the number of araC molecules bound, while j represents that of lacI. |
| - | According to [1], the reaction involved in the dynamics of the promotor state changes and reaction | + | According to [1], the reaction involved in the dynamics of the promotor state changes and the reaction can be summarized as follows:<BR> |
| - | < | + | [[Image:todai_susiki_matome1.jpg|400px|center]]<BR> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
where | where | ||
| - | + | <BR> | |
| - | < | + | [[Image:todai_susiki_matome2.jpg|200px|center]] |
| - | + | <BR> | |
| - | and X is the total number of ssrA tags in the system. k_{a}(It is a tag in order to quicken the disintegration process of the proteins.) The parameters of above reaction is listed as below: | + | and X is the total number of ssrA tags in the system. k_{a}(It is a tag in order to quicken the disintegration process of the proteins.) The parameters of above reaction is listed as below:<BR> |
| - | < | + | [[Image:todai_suusiki_matome3.jpg|400px|center]] |
===Modeling and the Result === | ===Modeling and the Result === | ||
| - | We set the initial state of the system to have only 75 | + | We set the initial state of the system to have only 75 [[Image:suusiki_last.jpg|50px]] and every other compound is set to zero. We used the SimBiology toolbox in MATLAB for the simulation, and the result is shown as the graph below. |
[[Image: Oscillation.png|500px]] | [[Image: Oscillation.png|500px]] | ||
| Line 118: | Line 87: | ||
The Lambda Phage switch forms the circuit by having double-stranded DNA. We borrowed a figure from [2] to show the complete structure of the switch. | The Lambda Phage switch forms the circuit by having double-stranded DNA. We borrowed a figure from [2] to show the complete structure of the switch. | ||
| - | [[Image: Lambda_switch_structure. | + | [[Image: Lambda_switch_structure.jpg|400px]] |
| - | From this figure we learnt that there are several independent promotor sites on the DNA that direction of transcription is not united. At the initial state, the system is filled with exceeding amount of cI. Therefore, promotors are repressive and the "Lambda switch" is turned off. When the system is exposed to UV light cI begin to disintegrate, as a result, the Lambda switch promotors are "turned on", and cro expression begin. This will facilitate the transcription on PL that makes N expresses. N has a NUT side upstream, which will quicken the transcription when N is bound, hence it forms a self-activate structure which could rapidly produces N. It will then help the expression of cII | + | From this figure we learnt that there are several independent promotor sites on the DNA that direction of transcription is not united. At the initial state, the system is filled with exceeding amount of cI. Therefore, promotors are repressive and the "Lambda switch" is turned off. When the system is exposed to UV light cI begin to disintegrate, as a result, the Lambda switch promotors are "turned on", and cro expression begin. This will facilitate the transcription on PL that makes N expresses. N has a NUT side upstream, which will quicken the transcription when N is bound, hence it forms a self-activate structure which could rapidly produces N. It will then help the expression of cII. In our model, a PlacI/araC promotor is inserted upstream of the cIII site. Thus, the switch alone could only express some amount of cII, which will not be enough to turn [[Image:todai_suuski_7.jpg|50px]] fully active. Therefore the system will end here, not being able to go back to the initial state. |
| + | In the model, we will be simplify the system by assuming the propriety of promotor [[Image:suusiki_6.jpg|50px]] and [[Image:todai_suuski_7.jpg|50px]] are the same, while [[Image:todai_suuski_8.jpg|50px]] and [[Image:todai_suuski_9.jpg|50px]]acts alike, the inference of cI to the [[Image:todai_suuski_9.jpg|50px]] promotor can be ignored. We further assume the NUT sites could be turned on when there are more RNAP.N.DNA than RNAP.DNA in the system. | ||
===Modeling and Result=== | ===Modeling and Result=== | ||
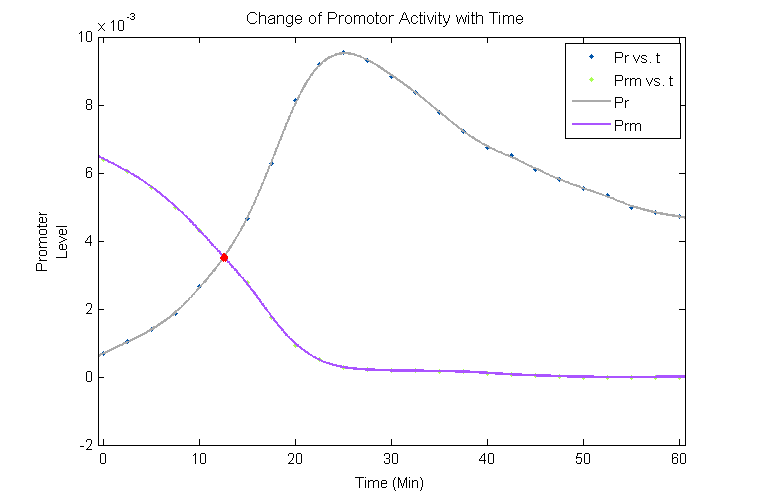
| - | According to [2] and [3], | + | According to [2] and [3], we tried to find the functional relationship between compounds and promotors, by fitting the time and expression of cro, cI and Promotor active level in Shea & Acker figure4 so that a mutual relationship among them could be examined. The curves gain by fitting the data are shown in the following graph. |
| + | |||
| + | [[Image: PR-PRM.png|500px]][[Image: CIcro.png|500px]] | ||
| + | |||
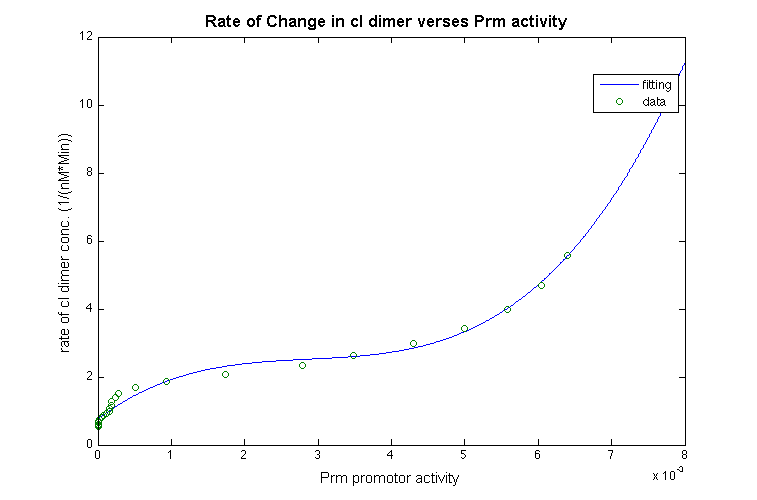
| + | With those approximation, we take the derivatives of the function for cI- dimer, and plot the data to estimate the relationship between the promotor and change in rate of the cI dimer shown below: | ||
| + | |||
| + | [[Image: Prm_DcI.png|500px]][[Image: Pr_Dcro.png|500px]] | ||
| + | |||
| + | |||
| + | Besides those functions, the rest of the reactions can be concluded as the following:<BR> | ||
| + | [[Image:Todai_suusiki_matome4.jpg|400px|center]] | ||
| + | |||
| + | where the parameters are indicated as following:<BR> | ||
| + | [[Image:Todai_suusiki_matome5.jpg|400px|center]] | ||
| + | |||
| + | |||
| + | We choose to have the initial value of cro and cI to be equal to the amount in the figure, while adjusting it with the Promotor-rate model after the PR promotor level reaches the peek. Once again with the help of MATLAB SimBiology toolbox, we could obtain the following trace regarding cI and cII. | ||
| + | |||
| + | [[Image: LambdaPhageSwitch.png|500px]] | ||
| + | |||
| + | As we can see from the graph, the cI concentration decay due to the UV light exposure, and cII could only have fractional amount in the system due to the concentration process. And cI could grow slowly, for the existence of [[Image:suusiki_6.jpg|50px]]. All in all, when the switch tested alone, it behave as a normal one-way switch. | ||
| + | =='''Integrated System'''== | ||
| + | As we stated in the overview for this Modeling, an integrated bioclock is the goal to our project. By having the discussion above, it become rather easy to combine the system. We consider the lacI site inserted after the cI site in Lambda phage switch. Due to our time scale is set to minutes, we will ignore the time delay between the transcription of cI and lacI. We then take promotor before cIII site into consideration that cIII could occupy P1 and P2 to lower the rate in which cII is concentrated. This would makes it possible for the promotor level of [[Image:todai_suuski_7.jpg|50px]] become high enough to begin effectively producing cI along with lacI. The system will eventually get exceeding amount of cI again, which should effectively "turn off" the switch by itself, going back to the initial state. After its whole process, we could take advantage of the time delay between the "on" and "off", therefore making the oscillation both controllable from outside and will stop according to its inside condition. | ||
| + | Unfortunately, we wasn't able to complete the simulation and present you with the actual data for the integrated system. However, we will try to finish it by Jamboree, | ||
| + | and should be able to show case the final data for the system as a whole. | ||
=='''Reference'''== | =='''Reference'''== | ||
Latest revision as of 02:26, 22 October 2009
| Home | The Team | The Project | Parts Submitted to the Registry | Modeling | Notebook | Protocols | Ethics |
|---|
Contents |
Overview
As we described in the project overview of the bioclock project, the actual clock consists of two separable parts: a bio-oscillator and a lambda phage switch. In this section we will be modeling each of the parts and have come up with an integrated model, resembling the ultimate goal of the project. By making comparisons between the results of our model and the results of the original article which we based our simulation upon, we examined how realistic the model is, and tried to gain a better understanding of how the oscillation process and, eventually, how the integrated model works.
Bio-Oscillator
Construction of the Model
In the natural world, there are quite a few systems that are known to act as an oscillator. The project started out by examining the behavior of Cyanobacteria. We learnt that the mutual activation and repression between several compounds resulted in the oscillation pattern of protein expression. We finally settled on creating a system utilizing lacI and araC as the mutual-interaction compound to construct a oscillator.
Although it is possible to trace the reactions in detail all the way from the transcription of mRNA and translation of the proteins with the precise reaction rates, it will be redundant to do so. In order to maintain clarity, we simplify this process to focus only on the rate of translation. One of the defining characteristics is the dual role of the promotor. It could be bound to a maximum of two lacI proteins and one araC protien. When araC is bound to it, it activates the translation of both araC and lacI, increasing lacI concentration while enhancing the possibility of lacI binding to the promotor as well. When the promotor is bound to lacI, it could transmute in to a looped form, to which araC cannot bind. We will note the state of promotor as ![]() , where i represents the number of araC molecules bound, while j represents that of lacI.
, where i represents the number of araC molecules bound, while j represents that of lacI.
According to [1], the reaction involved in the dynamics of the promotor state changes and the reaction can be summarized as follows:
where
and X is the total number of ssrA tags in the system. k_{a}(It is a tag in order to quicken the disintegration process of the proteins.) The parameters of above reaction is listed as below:
Modeling and the Result
We set the initial state of the system to have only 75 ![]() and every other compound is set to zero. We used the SimBiology toolbox in MATLAB for the simulation, and the result is shown as the graph below.
and every other compound is set to zero. We used the SimBiology toolbox in MATLAB for the simulation, and the result is shown as the graph below.
From the graph we could see the oscillation pattern stabilized after the first peek. And the period of oscillation is around 50 min. Since this is the result of a deterministic method, it is no surprise that the peeks are perfectly identical. From the result we get, it is safe to predict that the actual system should exhibit the oscillation pattern similar to the graph above with some possible noise and unstable behavior.
Lambda Phage Switch
What differentiate our bioclock from other similar system is we make it controllable from the outside world. The lambda pahge switch is a complex bio-system on its own, by twisting the circuit a little, adding a lacI coding site in detail, enables us to adapt the switch into the system, to get an integrated system which is exactly what iGEM is all about. In this section we will model the switching device and study its properties in depth.
Construction of the Model
The Lambda Phage switch forms the circuit by having double-stranded DNA. We borrowed a figure from [2] to show the complete structure of the switch.
From this figure we learnt that there are several independent promotor sites on the DNA that direction of transcription is not united. At the initial state, the system is filled with exceeding amount of cI. Therefore, promotors are repressive and the "Lambda switch" is turned off. When the system is exposed to UV light cI begin to disintegrate, as a result, the Lambda switch promotors are "turned on", and cro expression begin. This will facilitate the transcription on PL that makes N expresses. N has a NUT side upstream, which will quicken the transcription when N is bound, hence it forms a self-activate structure which could rapidly produces N. It will then help the expression of cII. In our model, a PlacI/araC promotor is inserted upstream of the cIII site. Thus, the switch alone could only express some amount of cII, which will not be enough to turn ![]() fully active. Therefore the system will end here, not being able to go back to the initial state.
In the model, we will be simplify the system by assuming the propriety of promotor
fully active. Therefore the system will end here, not being able to go back to the initial state.
In the model, we will be simplify the system by assuming the propriety of promotor ![]() and
and ![]() are the same, while
are the same, while ![]() and
and ![]() acts alike, the inference of cI to the
acts alike, the inference of cI to the ![]() promotor can be ignored. We further assume the NUT sites could be turned on when there are more RNAP.N.DNA than RNAP.DNA in the system.
promotor can be ignored. We further assume the NUT sites could be turned on when there are more RNAP.N.DNA than RNAP.DNA in the system.
Modeling and Result
According to [2] and [3], we tried to find the functional relationship between compounds and promotors, by fitting the time and expression of cro, cI and Promotor active level in Shea & Acker figure4 so that a mutual relationship among them could be examined. The curves gain by fitting the data are shown in the following graph.
With those approximation, we take the derivatives of the function for cI- dimer, and plot the data to estimate the relationship between the promotor and change in rate of the cI dimer shown below:
Besides those functions, the rest of the reactions can be concluded as the following:
where the parameters are indicated as following:
We choose to have the initial value of cro and cI to be equal to the amount in the figure, while adjusting it with the Promotor-rate model after the PR promotor level reaches the peek. Once again with the help of MATLAB SimBiology toolbox, we could obtain the following trace regarding cI and cII.
As we can see from the graph, the cI concentration decay due to the UV light exposure, and cII could only have fractional amount in the system due to the concentration process. And cI could grow slowly, for the existence of ![]() . All in all, when the switch tested alone, it behave as a normal one-way switch.
. All in all, when the switch tested alone, it behave as a normal one-way switch.
Integrated System
As we stated in the overview for this Modeling, an integrated bioclock is the goal to our project. By having the discussion above, it become rather easy to combine the system. We consider the lacI site inserted after the cI site in Lambda phage switch. Due to our time scale is set to minutes, we will ignore the time delay between the transcription of cI and lacI. We then take promotor before cIII site into consideration that cIII could occupy P1 and P2 to lower the rate in which cII is concentrated. This would makes it possible for the promotor level of ![]() become high enough to begin effectively producing cI along with lacI. The system will eventually get exceeding amount of cI again, which should effectively "turn off" the switch by itself, going back to the initial state. After its whole process, we could take advantage of the time delay between the "on" and "off", therefore making the oscillation both controllable from outside and will stop according to its inside condition.
become high enough to begin effectively producing cI along with lacI. The system will eventually get exceeding amount of cI again, which should effectively "turn off" the switch by itself, going back to the initial state. After its whole process, we could take advantage of the time delay between the "on" and "off", therefore making the oscillation both controllable from outside and will stop according to its inside condition.
Unfortunately, we wasn't able to complete the simulation and present you with the actual data for the integrated system. However, we will try to finish it by Jamboree, and should be able to show case the final data for the system as a whole.
Reference
[1] Jesse Stricker, Scott Cookson, Matthew R. Bennett, William H. Mather, Lev S. Tsimring and Jeff Hasty, 2008 A fast, robust and tunable synthetic gene oscillator ( and Supplementary Information) Nature 07389 Vol 456: 516-519
[2] Adam Arkin, John Ross and Harley H. McAdams, 1998 Stochastic Kinetic Analysis of Developmental Pathway Bifurcation in Phage lambda-Infected Escherichia coli Cells Genetics 149: 1633–1648
[3] Shea, M. A., and G. K. Ackers, 1985 The OR control system of bacteriophage lambda: a physical-chemical model for gene regulation J. Mol. Biol. 181: 211–230
 "
"