Team:Bologna
From 2009.igem.org
Marco.cavina (Talk | contribs) |
Marco.cavina (Talk | contribs) |
||
| (78 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | < | + | {{Template:BolognaTemplate}} |
| - | <style | + | <br><br><br> |
| - | + | [[Image:Ely9Copia.jpg|center|850px]] | |
| - | + | <br> | |
| + | <div style="text-align:justify"> | ||
| - | + | = Project Summary = | |
| - | + | ||
| - | + | <br> | |
| - | + | <font face="Calibri" size="5"> | |
| - | + | '''Our idea''' | |
| - | + | </font> | |
| - | + | <br><br> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <html> | |
| + | <font face="Calibri" font size="4" color="#000000"> | ||
| + | The aim of our project is the design of a standard device to control the synthesis of any protein of interest. This "general-purpose" device, implemented in <i>E. coli</i>, acts at the translational level to allow silencing of protein expression faster than using regulated promoters. We named this device <b>T-REX</b> (<b>T</b>rans <b>R</b>epressor of <b>Ex</b>pression). | ||
| + | </font></html> | ||
| + | <br> | ||
| + | <font face="Calibri" size="5"> | ||
| + | '''How T-REX works''' | ||
| + | </font> | ||
| + | <br><br> | ||
| - | |||
| - | < | + | <font face="Calibri" font size="4" color="#000000"> |
| - | = | + | <html> |
| + | The device consists of two new BioBricks: | ||
| + | <br> | ||
| + | <ul> | ||
| + | <li><font color="#000080"><b>CIS-repressing</b></font>, to be assembled upstream of the target coding sequence. | ||
| + | </ul> | ||
| + | <ul> | ||
| + | <li><font color="#000080"><b>TRANS-repressor</b></font>, complementary to the CIS-repressing and placed under the control of a different promoter. | ||
| + | </ul></html> | ||
| + | <br> | ||
| + | CIS-repressing and TRANS-repressor sequences were designed by [[Team:Bologna/Software#1|BASER]] software. | ||
| + | <br><br> | ||
| + | Transcription of the target gene yields a mRNA strand - containing the CIS-repressing sequence at its 5' end - available for translation into protein by ribosomes (<i>see Fig. 1, left panel</i>). When the promoter controlling the TRANS coding sequence is active, it drives the transcription of an oligoribonucleotide complementary to the CIS mRNA sequence. The TRANS/CIS <b>RNA duplex</b> prevents ribosomes from binding to RBS on target mRNA, thus <b>silencing protein synthesis</b>. The amount of the TRANS-repressor regulates the rate of translation of the target mRNA (<i>see Fig. 1, right panel</i>) | ||
| + | <br><br> | ||
| + | [[Image:project3b.png|center|950px|thumb|<center>Figure 1 - T-REX device</center>]] | ||
| + | </font> | ||
| - | ''' | + | <br><br> |
| + | <font face="Calibri" size="5"> | ||
| + | '''How we can test the device''' | ||
| + | </font> | ||
| + | <br><br><br> | ||
| + | <html> | ||
| + | <font face="Calibri" font size="4" color="#000000"> | ||
| + | In order to test and characterize our T-REX device, we developed the following genetic circuit (Fig 2): | ||
| + | </font></html> | ||
| + | <br><br> | ||
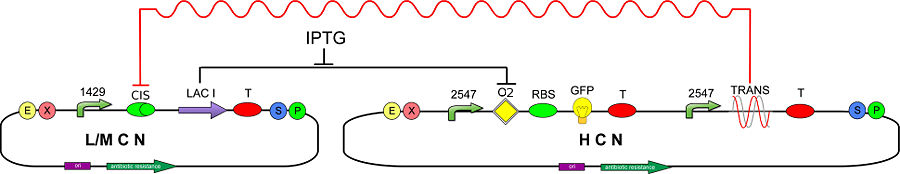
| + | [[Image:circuit2OK.jpg|center|900px|thumb|<center>Figure 2 - Genetic Circuit to test CIS and TRANS' mRNA affinity</center>]] | ||
| + | <br><br> | ||
| + | <br> | ||
| + | <font size="4"> | ||
| + | More details about our work are reported in the [[Team:Bologna/Project|Project]] section. | ||
| + | </font> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| - | + | = Acknowledgements = | |
| + | <br> | ||
| + | <font size="3"> | ||
| + | * ''' [http://www.unibo.it/Portale/default.htm University of Bologna] ''' | ||
| + | <br> | ||
| + | [[Image:LogoUnibo.jpg|left|100 px]] | ||
| - | + | <br><br><br><br><br><br><br> | |
| - | + | * ''' [http://serinar.criad.unibo.it Ser.In.Ar. Cesena] ''' | |
| - | + | ||
| + | <br> | ||
| + | [[Image:Ser_In_Ar.jpg|left|500px]] | ||
| - | + | <br><br><br><br><br> | |
| + | * <font color=#0000cd>'''Cultural Association San Sebastiano'''</font> | ||
| - | + | [[Image:SSebastiano.jpg|left|200px]] | |
| - | + | <br><br><br><br><br><br><br><br> | |
| - | + | ---- | |
| - | |||
| - | |||
| - | |||
<html> | <html> | ||
| - | < | + | <center> |
| - | / | + | <a href="http://www2.clustrmaps.com/counter/maps.php?url=https://2009.igem.org/Team:Bologna" id="clustrMapsLink"><img src="http://www2.clustrmaps.com/counter/index2.php?url=https://2009.igem.org/Team:Bologna" style="border:0px;" alt="Locations of visitors to this page" title="Locations of visitors to this page" id="clustrMapsImg" onerror="this.onerror=null; this.src='http://clustrmaps.com/images/clustrmaps-back-soon.jpg'; document.getElementById('clustrMapsLink').href='http://clustrmaps.com';" /> |
| - | + | </a> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | </center> | |
| - | + | </html> | |
| - | + | ||
| - | |||
| - | |||
| - | + | [https://2009.igem.org/Team:Bologna ''Up''] | |
| - | + | </font> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | </ | + | |
Latest revision as of 03:12, 22 October 2009
| HOME | TEAM | PROJECT | SOFTWARE | MODELING | WET LAB | PARTS | HUMAN PRACTICE | JUDGING CRITERIA |
|---|
Project Summary
Our idea
The aim of our project is the design of a standard device to control the synthesis of any protein of interest. This "general-purpose" device, implemented in E. coli, acts at the translational level to allow silencing of protein expression faster than using regulated promoters. We named this device T-REX (Trans Repressor of Expression).
How T-REX works
The device consists of two new BioBricks:
CIS-repressing and TRANS-repressor sequences were designed by BASER software.
Transcription of the target gene yields a mRNA strand - containing the CIS-repressing sequence at its 5' end - available for translation into protein by ribosomes (see Fig. 1, left panel). When the promoter controlling the TRANS coding sequence is active, it drives the transcription of an oligoribonucleotide complementary to the CIS mRNA sequence. The TRANS/CIS RNA duplex prevents ribosomes from binding to RBS on target mRNA, thus silencing protein synthesis. The amount of the TRANS-repressor regulates the rate of translation of the target mRNA (see Fig. 1, right panel)
How we can test the device
In order to test and characterize our T-REX device, we developed the following genetic circuit (Fig 2):
More details about our work are reported in the Project section.
Acknowledgements
- [http://www.unibo.it/Portale/default.htm University of Bologna]
- [http://serinar.criad.unibo.it Ser.In.Ar. Cesena]
- Cultural Association San Sebastiano
 "
"