Team:UNICAMP-Brazil/Coliguard/Recognition
From 2009.igem.org
(→Recognition by conjugation) |
(→Py promoter cloning and characterization) |
||
| (15 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:UNICAMP-Brazil/inc_topo}} | {{:Team:UNICAMP-Brazil/inc_topo}} | ||
| + | __NOTOC__ | ||
=The Coliguard - Recognition= | =The Coliguard - Recognition= | ||
| Line 6: | Line 7: | ||
We aim to create bacteria that can protect themselves against contaminants. So, our bacteria need to be able to kill other organisms which may appear in the culture medium. | We aim to create bacteria that can protect themselves against contaminants. So, our bacteria need to be able to kill other organisms which may appear in the culture medium. | ||
| - | + | ||
| + | *Why not create a continuous mechanism of eliminating contaminants? | ||
A simple way to do this would be to create bacteria that express a toxin constitutively. However, if the bacteria produce the toxin continuously, they will compromise their metabolic energy for the production of the toxic compound and, in addition, the production of large amounts of these antibiotic substances may contribute to the problem of antibiotic resistance. Moreover, the toxin itself would be a type of contaminant in the industrial process, requiring resources to be allocated in order to remove the toxin from the medium and purify the compound of interest. | A simple way to do this would be to create bacteria that express a toxin constitutively. However, if the bacteria produce the toxin continuously, they will compromise their metabolic energy for the production of the toxic compound and, in addition, the production of large amounts of these antibiotic substances may contribute to the problem of antibiotic resistance. Moreover, the toxin itself would be a type of contaminant in the industrial process, requiring resources to be allocated in order to remove the toxin from the medium and purify the compound of interest. | ||
| - | + | *Therefore, we need a contaminant-sensitive recognition system! | |
We need to create a contaminant-sensitive mechanism that will allow the detection of the contaminants and trigger the killing mechanism. In this case, the toxin will be produced only when the contaminants are present. | We need to create a contaminant-sensitive mechanism that will allow the detection of the contaminants and trigger the killing mechanism. In this case, the toxin will be produced only when the contaminants are present. | ||
| - | + | *How will our bacterial guards recognize the contaminants? | |
Our idea is based on the premise that the engineered ''E. coli'' must be able to recognize contaminants in the culture medium as non-self. Thus, our bacteria need to recognize a specific signal that they don’t produce, but which are broadly produced or presented by potential contaminants. Thinking about the potential signals, we found two candidates: the AI2 (auto inducer 2) and the conjugation recognition mechanism. | Our idea is based on the premise that the engineered ''E. coli'' must be able to recognize contaminants in the culture medium as non-self. Thus, our bacteria need to recognize a specific signal that they don’t produce, but which are broadly produced or presented by potential contaminants. Thinking about the potential signals, we found two candidates: the AI2 (auto inducer 2) and the conjugation recognition mechanism. | ||
| - | + | *Using AI-2 as a signal. How? | |
As most bacterial species produces AI2 as a secondary metabolite(1; 2; 3), we decided to use this compound as a recognition factor. Our ''E. coli'' will be an AI2- strain and won´t produce native AI2. The presence of AI2 in the culture medium will then indicate the presence of contaminants, which will be recognized by an AI2 sensitive promoter present in our ''E. coli''. This promoter will control the differentiation and killing mechanisms (see the links for more information). | As most bacterial species produces AI2 as a secondary metabolite(1; 2; 3), we decided to use this compound as a recognition factor. Our ''E. coli'' will be an AI2- strain and won´t produce native AI2. The presence of AI2 in the culture medium will then indicate the presence of contaminants, which will be recognized by an AI2 sensitive promoter present in our ''E. coli''. This promoter will control the differentiation and killing mechanisms (see the links for more information). | ||
| - | + | *But, what if the contaminants do not produce AI-2? | |
In these situations, our ''E.coli'' will use another recognition mechanism, based in conjugation. In conjugation, a donor bacterium only conjugates with organisms that don’t have the same conjugative plasmid. Bacteria that already have the plasmid display membrane proteins that prevent the conjugation with other bacteria that have the same conjugative plasmid (4). Therefore, if our ''E.coli'' has a conjugative plasmid, they will only conjugate with different organisms, which will be the contaminants. | In these situations, our ''E.coli'' will use another recognition mechanism, based in conjugation. In conjugation, a donor bacterium only conjugates with organisms that don’t have the same conjugative plasmid. Bacteria that already have the plasmid display membrane proteins that prevent the conjugation with other bacteria that have the same conjugative plasmid (4). Therefore, if our ''E.coli'' has a conjugative plasmid, they will only conjugate with different organisms, which will be the contaminants. | ||
| Line 39: | Line 41: | ||
Combining these two recognition systems, our ''E. coli'' will be able to recognize a vast group of contaminants. | Combining these two recognition systems, our ''E. coli'' will be able to recognize a vast group of contaminants. | ||
| + | |||
==Recognition by conjugation== | ==Recognition by conjugation== | ||
| Line 46: | Line 49: | ||
Our idea is to use the ''Py'' promoter to act as a second signal (in addition to AI-2) to recognize contaminants, controlling the expression of the ''AI-2'' gene and, consequently, the killing and the differentiation mechanisms that are AI-2-dependent. In order to do that, we first need to isolate the promoter and then, characterize its operation. | Our idea is to use the ''Py'' promoter to act as a second signal (in addition to AI-2) to recognize contaminants, controlling the expression of the ''AI-2'' gene and, consequently, the killing and the differentiation mechanisms that are AI-2-dependent. In order to do that, we first need to isolate the promoter and then, characterize its operation. | ||
In parallel, we will improve F plasmid to use in our engineered ''E. coli'' by, removing uninteresting parts in order to make it smaller and easier to copy. Moreover, we will remove the insertion sequences, which are the portions of the F plasmid DNA that mediate the integration into the bacterial chromosome, thus avoiding possible problems caused by F plasmid integration. (7). Moreover, we will add different replication origins to the plasmid, thus increasing the range of contaminants that may be recognized (8). | In parallel, we will improve F plasmid to use in our engineered ''E. coli'' by, removing uninteresting parts in order to make it smaller and easier to copy. Moreover, we will remove the insertion sequences, which are the portions of the F plasmid DNA that mediate the integration into the bacterial chromosome, thus avoiding possible problems caused by F plasmid integration. (7). Moreover, we will add different replication origins to the plasmid, thus increasing the range of contaminants that may be recognized (8). | ||
| + | |||
==Py promoter isolation== | ==Py promoter isolation== | ||
| Line 54: | Line 58: | ||
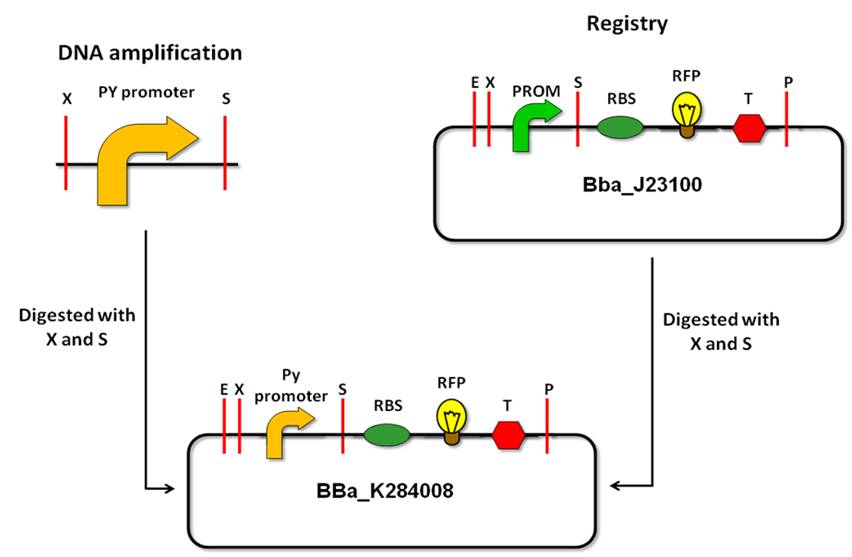
The strategy consists of PCR amplification of the ''Py'' promoter region by using primers that contain Biobrick prefix and Biobrick suffix, thus allowing us to clone the amplicon as Biobrick parts. Before designing primers, we verified that none of the restriction sites of the prefix or sufixe are contained in the amplified sequence. | The strategy consists of PCR amplification of the ''Py'' promoter region by using primers that contain Biobrick prefix and Biobrick suffix, thus allowing us to clone the amplicon as Biobrick parts. Before designing primers, we verified that none of the restriction sites of the prefix or sufixe are contained in the amplified sequence. | ||
| - | What we have done | + | '''What we have done''' |
| - | We designed primers from the F plasmid sequence ( | + | We designed primers from the F plasmid sequence (5) to isolate the ''Py'' promoter by PCR. We used information on scientific literature to determine the region of the promoter (5; 6). We found that there is an intrinsic curvature, upstream of the promoter region, which could interfere in the promoter regulation. We decided, therefore, to design two pairs of primers: PyF1 - PyR1 and PyF2 - PyR1. These primer sets have the same reverse primers but different forward primers. The PyF1 – PyR1 amplicon has 133bp and includes the two closer bends upstream of the promoter region. The PyF2 - PyR1 amplicon has 75bp and doesn’t include any bend (see Notebooks – September 4th for more details). |
We managed to amplify the pY promoter amplicons through PCR from the E. coli F plasmid, creating two new Biobrick parts: BBa_K284007 (PyF2 - PyR1 amplicon) and BBa_K284008 (PyF1 - PyR1 amplicon). | We managed to amplify the pY promoter amplicons through PCR from the E. coli F plasmid, creating two new Biobrick parts: BBa_K284007 (PyF2 - PyR1 amplicon) and BBa_K284008 (PyF1 - PyR1 amplicon). | ||
| - | == | + | ==Py promoter cloning and characterization== |
'''Strategy''' | '''Strategy''' | ||
| - | After isolation of the ''Py'' promoter, it must be cloned in the BioBrick vector according to the standard assembly strategy. Afterwards, to characterize the promoter activity, we will make a device, with a reporter gene under the control of the ''Py'' promoter. We decided to use a fluorescent protein as a reporter gene for this characterization, such as the GFP (Green Fluorescent Protein). This reporter system will allow us to use the fluorescence signal in order to quantify promoter activity in a spectrofluorometer. ( | + | After isolation of the ''Py'' promoter, it must be cloned in the BioBrick vector according to the standard assembly strategy. Afterwards, to characterize the promoter activity, we will make a device, with a reporter gene under the control of the ''Py'' promoter. We decided to use a fluorescent protein as a reporter gene for this characterization, such as the GFP (Green Fluorescent Protein). This reporter system will allow us to use the fluorescence signal in order to quantify promoter activity in a spectrofluorometer (Hitachi F-4500). |
| + | The characterization experiment consists of a conjugation experiment (9) to be conducted using our engineered ''E. coli'' as the donor bacterium and another ''E. coli'', without the F plasmid and containing a marker of resistance different from the F plasmid, as the receiver bacterium. After the beginning of the conjugation process, we will take samples after 1h, 2h, 3h, 6h, 9h, 12h and 24h. We will analyze these samples in a spectrofluorometer to quantify the fluorescence emitted. In addition, we will inoculate a portion of the sample in Petri dishes containing LB medium with the appropriated antibiotics to select only the receiver bacteria with the F plasmid. The plates will be incubated overnight at 37°C and then we will quantify the conjugation events through the observation of bacteria growing in the plates. Through this experiment, we will be able to analyze the ''Py'' promoter activity data in comparison with the conjugation events. If the measured Py promoter activity and the conjugation process begin coincide, we may conclude that the experiment worked and that the ''Py'' promoter can be used as a signal for conjugation and could thus be adapted into our recognition mechanism. | ||
| - | + | Literature reports that the ''Py'' promoter has a basal activity (5; 6). However, this fact won’t be a problem because its function will be adapted to the control of the production of AI-2 in the presence of contaminants. AI-2 is a type of signal that needs to exceed a threshold in order to stimulate the promoter sensitive to AI-2 (10), which will be in turn coupled to the differentiation and the killing mechanisms. Therefore, the basal activity of the ''Py'' promoter should not be enough to trigger the differentiation and killing mechanisms. | |
| - | + | '''What we have done''' | |
| - | + | ||
| - | What we have done | + | |
At first we intended to use the GFP as a reporter gene. We took the BBa_E0040 part, which contains the GFP, and started to work with it. Unfortunately, it didn’t work as expected. We did some restriction analyses and the bands expected after the electrophoresis running didn’t appear. | At first we intended to use the GFP as a reporter gene. We took the BBa_E0040 part, which contains the GFP, and started to work with it. Unfortunately, it didn’t work as expected. We did some restriction analyses and the bands expected after the electrophoresis running didn’t appear. | ||
| Line 79: | Line 82: | ||
| - | In the characterization experiment, we will use a donor bacterium that carries an F plasmid with the ampicillin resistance gene. Thus, we can’t select transformed bacterial colonies with our device in a medium supplemented with ampicillin. Therefore, we cloned the BBa_K284008 part into the pSB1AK3 vector, which carries the ampicillin and kanamycin resistance genes. Then, we could select the colonies in a medium supplemented with kanamycin. | + | In the characterization experiment, we will use a donor bacterium that carries an F plasmid with the ampicillin resistance gene. Thus, we can’t select transformed bacterial colonies with our device in a medium supplemented with ampicillin. Therefore, we cloned the BBa_K284008 part into the pSB1AK3 vector, which carries the ampicillin and kanamycin resistance genes. Then, we could select the colonies in a medium supplemented with kanamycin. |
==Engineering the F plasmid – removing parts and adding new replication origins== | ==Engineering the F plasmid – removing parts and adding new replication origins== | ||
| Line 85: | Line 88: | ||
'''Strategy''' | '''Strategy''' | ||
| - | First of all, we will reduce the size of the F plasmid by removing uninteresting parts and the insertion sequences, which are the portions of the F plasmid DNA that mediate the integration into the bacterial chromosome (generating Hfr strains). As Willetts and Johnson reported ( | + | First of all, we will reduce the size of the F plasmid by removing uninteresting parts and the insertion sequences, which are the portions of the F plasmid DNA that mediate the integration into the bacterial chromosome (generating Hfr strains). As Willetts and Johnson reported (7), we will remove a part from the F plasmid by single digestion with HindIII followed by recircularization. This process creates pED100, a conjugative F plasmid derivative without insertion sequences. Through this simple procedure we will reduce considerably the possibility of plasmid integration into the chromosome. |
| - | After that, we will add multiple replication origins to our F plasmid. In consequence, we will reduce conjugation specificity, thus increasing the range of contaminant recognition | + | After that, we will add multiple replication origins to our F plasmid (8). In consequence, we will reduce conjugation specificity, thus increasing the range of contaminant recognition.Our strategy is to use the replication origins of plasmids from different groups of bacteria, in an effort to reduce as much as we can the conjugation specificity. |
| - | What we have done | + | '''What we have done''' |
| - | We managed to remove the insertions sequences and the uninteresting parts from the F plasmid. At this way, our plasmid is now easier to copy and to work with. | + | We managed to remove the insertions sequences and the uninteresting parts from the F plasmid. At this way, our plasmid is now easier to copy and to work with. |
| + | |||
==The recognition by AI-2== | ==The recognition by AI-2== | ||
| Line 101: | Line 105: | ||
Therefore, we need a mechanism in which both recognition systems can be detected. In order to achieve that, the production of AI-2 has to be controlled by ''Py'' promoter. The LuxS gene, which encodes for the LuxS synthase, produce DPD (4,5-dihydroxy-2,3-pentanedione) that spontaneously forms the AI-2 molecule. This part was already designed by Davidson Missouri Western in 2008 and we will use it to produce AI-2 under the control of the ''Py'' promoter. In case of conjugation, ''Py'' will be activated and the LuxS synthase will be produced. On the other hand, microorganisms that naturally produce the AI-2 molecule skip that step. Once the recognition is achieved, it triggers the differentiation process through the expression of the CRE-recombinase. The Cre-recombinase will be under the control of the ''QseA'' promotor, which is activated in the presence of AI-2. The ''QseA'' also is known as the quorum-sensing ''E. coli'' regulator A and an array study showed that this regulator is regulated by AI-2 (10). The CRE-recombinase was already designed by Arkin Lab and will be utlized in this project. | Therefore, we need a mechanism in which both recognition systems can be detected. In order to achieve that, the production of AI-2 has to be controlled by ''Py'' promoter. The LuxS gene, which encodes for the LuxS synthase, produce DPD (4,5-dihydroxy-2,3-pentanedione) that spontaneously forms the AI-2 molecule. This part was already designed by Davidson Missouri Western in 2008 and we will use it to produce AI-2 under the control of the ''Py'' promoter. In case of conjugation, ''Py'' will be activated and the LuxS synthase will be produced. On the other hand, microorganisms that naturally produce the AI-2 molecule skip that step. Once the recognition is achieved, it triggers the differentiation process through the expression of the CRE-recombinase. The Cre-recombinase will be under the control of the ''QseA'' promotor, which is activated in the presence of AI-2. The ''QseA'' also is known as the quorum-sensing ''E. coli'' regulator A and an array study showed that this regulator is regulated by AI-2 (10). The CRE-recombinase was already designed by Arkin Lab and will be utlized in this project. | ||
| - | Constructions: | + | '''Constructions:''' |
| - | 1) BBa_K284027 - ''QseA'' promoter | + | '''1)''' BBa_K284027 - ''QseA'' promoter |
This part will be constructed using the standard assembly strategy. After the amplification of the ''QseA'' promoter the part will be joined with the Bba_J23100 plasmid for characterization. | This part will be constructed using the standard assembly strategy. After the amplification of the ''QseA'' promoter the part will be joined with the Bba_J23100 plasmid for characterization. | ||
| Line 110: | Line 114: | ||
| - | 2) BBa_K284028 – LuxS under the control of ''Py'' promoter | + | '''2)''' BBa_K284028 – LuxS under the control of ''Py'' promoter |
This device will be composed of BBa_K284008 (Py promoter), BBa_B0034 (a RBS), BBa_K091109 (LuxS) and BBa_0014 (a double terminator) and constructed following the standard assembly strategy. | This device will be composed of BBa_K284008 (Py promoter), BBa_B0034 (a RBS), BBa_K091109 (LuxS) and BBa_0014 (a double terminator) and constructed following the standard assembly strategy. | ||
| Line 116: | Line 120: | ||
[[Image:Imagem3.jpg|center|400px]] | [[Image:Imagem3.jpg|center|400px]] | ||
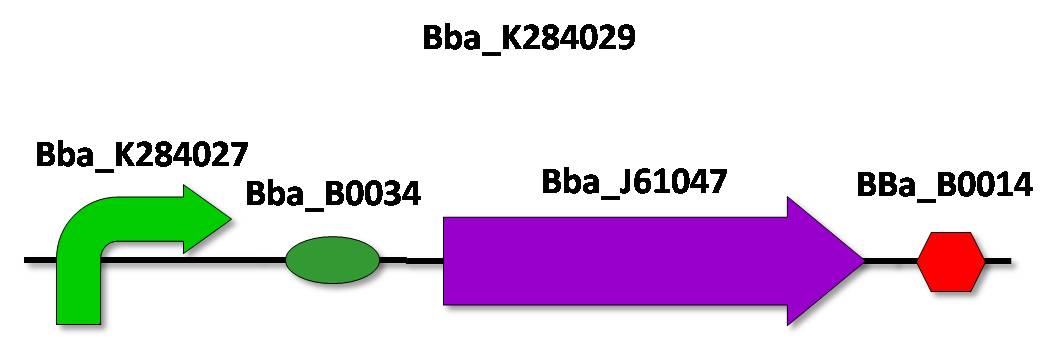
| - | 3) BBa_K284029 – CRE-recombinase under the control of ''QseA'' promoter | + | '''3)''' BBa_K284029 – CRE-recombinase under the control of ''QseA'' promoter |
This device will be composed of BBa_K28402 (QseA promoter), BBa_B0034 (a RBS), BBa_J61047 (CRE-recombinase) and BBa_0014 (a double terminator) and constructed according to the standard assembly strategy. | This device will be composed of BBa_K28402 (QseA promoter), BBa_B0034 (a RBS), BBa_J61047 (CRE-recombinase) and BBa_0014 (a double terminator) and constructed according to the standard assembly strategy. | ||
| Line 123: | Line 127: | ||
[[Image:Imagem4.jpg|center|400px]] | [[Image:Imagem4.jpg|center|400px]] | ||
| - | ==References== | + | |
| + | ===References=== | ||
| + | ---- | ||
1. Cao, J.G. & Meighen, E.A. J Biol Chem. 1989, 264, 21670-21676. | 1. Cao, J.G. & Meighen, E.A. J Biol Chem. 1989, 264, 21670-21676. | ||
Latest revision as of 03:17, 22 October 2009
| ||||||||||||||||||||||||||||||||||
 "
"