Team:Nevada/Project
From 2009.igem.org
(→Goal 3) |
(→Special Acknowledgements) |
||
| (46 intermediate revisions not shown) | |||
| Line 116: | Line 116: | ||
== '''Overall project''' == | == '''Overall project''' == | ||
[[Image:iGEMlogo31-1.jpg|none|frame|alt=alt text|Simplified Graphic of our project goal]] | [[Image:iGEMlogo31-1.jpg|none|frame|alt=alt text|Simplified Graphic of our project goal]] | ||
| - | |||
'''Cinnamicide: Producing a Natural Insecticide against Mosquito Larvae in ''E. coli'' and Duckweed''' | '''Cinnamicide: Producing a Natural Insecticide against Mosquito Larvae in ''E. coli'' and Duckweed''' | ||
Cinnamaldehyde is a natural insecticide against mosquito larvae that shows low toxicity towards other organisms. The objective of this project is to engineer the cinnamaldehyde biosynthetic pathway into ''E. coli'' to develop an inexpensive and readily available source of this compound. By introducing the genes encoding phenylalanine-ammonia lyase (gene 1), 4-coumarate:CoA ligase (gene 2), and cinnamoyl-CoA reductase (gene 3), it should be possible to produce cinnamaldehyde from available phenylalanine in ''E. coli''. Once we have demonstrated that we can produce cinnamaldehyde in ''E. coli'', we will engineer cinnamaldehyde production in duckweed (''Wolffia'' or ''Lemna'' species), a small aquatic plant. Because mosquito larvae feed on duckweed detritus, the engineered plant will serve as an excellent vehicle to deliver cinnamaldehyde for mosquito control. | Cinnamaldehyde is a natural insecticide against mosquito larvae that shows low toxicity towards other organisms. The objective of this project is to engineer the cinnamaldehyde biosynthetic pathway into ''E. coli'' to develop an inexpensive and readily available source of this compound. By introducing the genes encoding phenylalanine-ammonia lyase (gene 1), 4-coumarate:CoA ligase (gene 2), and cinnamoyl-CoA reductase (gene 3), it should be possible to produce cinnamaldehyde from available phenylalanine in ''E. coli''. Once we have demonstrated that we can produce cinnamaldehyde in ''E. coli'', we will engineer cinnamaldehyde production in duckweed (''Wolffia'' or ''Lemna'' species), a small aquatic plant. Because mosquito larvae feed on duckweed detritus, the engineered plant will serve as an excellent vehicle to deliver cinnamaldehyde for mosquito control. | ||
| + | |||
==Safety Questionnaire== | ==Safety Questionnaire== | ||
| Line 131: | Line 131: | ||
<BR><BR> | <BR><BR> | ||
'''Is there a local biosafety group, committee, or review board at your institution?''' <BR> | '''Is there a local biosafety group, committee, or review board at your institution?''' <BR> | ||
| - | Yes, there is | + | Yes, there is an Institutional Biosafety Committee (IBC) at our institution which is part of the Environmental Health & Safety (EH&S) program. We also collaborated with Scott Munsen of the Washoe County Vector Control Program. |
<BR><BR> | <BR><BR> | ||
'''What does your local biosafety group think about your project?''' | '''What does your local biosafety group think about your project?''' | ||
<BR> | <BR> | ||
| - | Our local biosafety group approved our project design and subsequent facilitation of research. Scott Munsen of the Washoe County Vector Control believes this could be a | + | Our local biosafety group approved our project design and subsequent facilitation of research. Scott Munsen of the Washoe County Vector Control Program believes this could be a feasible pest control and will aid in the larvicidal tests of cinnamaldehyde against various mosquito species to verify its larvicidal activity and to ensure we are not harming non-target organisms. |
<BR><BR> | <BR><BR> | ||
'''Do any of the new BioBrick parts that you made this year raise any safety issues?''' | '''Do any of the new BioBrick parts that you made this year raise any safety issues?''' | ||
<BR> | <BR> | ||
| - | No, none of our new BioBrick parts that we made this year raise any safety issues. The genes we are using are naturally occurring genes expressed in plants and other organisms that do not | + | No, none of our new BioBrick parts that we made this year raise any safety issues. The genes we are using are naturally occurring genes expressed in plants and other organisms that do not pose any risk. |
==Project Details== | ==Project Details== | ||
| Line 145: | Line 145: | ||
=== '''Why Produce an Insecticide against Mosquitoes?''' === | === '''Why Produce an Insecticide against Mosquitoes?''' === | ||
[[Image:180px-Aedes_aegypti_biting_human.jpg|right|frame|alt=alt text|Aedes aegypti biting human]] | [[Image:180px-Aedes_aegypti_biting_human.jpg|right|frame|alt=alt text|Aedes aegypti biting human]] | ||
| - | It is estimated that every year over 350-500 million people are infected by diseases spread by mosquitoes, and over 1 million of them die as a result. Current insecticides being used which include dichlorodiphenyltrichloroethane (DDT), organophosphates, and others, cause environmental | + | It is estimated that every year over 350-500 million people are infected by diseases spread by mosquitoes, and over 1 million of them die as a result. Current insecticides being used which include dichlorodiphenyltrichloroethane (DDT), organophosphates, and others, cause environmental damage to non-target organisms, including humans. Many mosquito species gain resistance to these insecticides, so new ways to combat mosquitoes that transmit human diseases need to be produced. In order to replace these general synthetic insecticides, our proposed project is to produce a natural, low toxicity mosquito larvicide through the production of cinnamaldehyde. Finding a cost-effective, environmentally friendly manner to manage mosquito populations is a key aim of this project. |
=== Goal 1 === | === Goal 1 === | ||
| - | Develop a mathematical model to predict pathway flux based on enzyme kinetic values and | + | Develop a mathematical model to predict pathway flux based on enzyme kinetic values and levels of gene expression. The information obtained from this model will help rationalize engineering design choices to optimize cinnamaldehyde production in ''E.coli''. |
=== Experiment and Results for Goal 1=== | === Experiment and Results for Goal 1=== | ||
| - | 1. Using Mathematica, a model was developed to describe Michaelis-Menten steady-state kinetics for the engineered cinnamaldehyde biosynthetic pathway. The published Km and Vmax values for the plant | + | 1. Using Mathematica, a model was developed to describe Michaelis-Menten steady-state kinetics for the engineered cinnamaldehyde biosynthetic pathway. The published Km and Vmax values for the plant enzyme, phenylalanine-ammonia lyase, 4-coumarate:CoA ligase and cinnamoyl-CoA reductase (feruloyl-CoA reductase), were used to build our model. Using wild type kinetic parameters and the assumption that all three enzymes were present in equal amounts, the model predicted that a biosynthetic bottleneck will occur at 4-coumarate:CoA ligase. This problem appears to be primarily due to the high Km value of the enzyme for its substrate cinnamic acid. Therefore, we used the model to test two possible means of increasing pathway flux: 1) increasing 4-coumarate:CoA ligase levels and 2) decreasing 4-coumarate:CoA ligase’s Km for cinnamic acid. The results produced from the model showed that by increasing the 4-coumarate:CoA ligase concentration ten-fold relative to the other enzymes in the pathway, or by decreasing the Km forty-fold, the barrier could be bypassed in this way. the results of these studies are shown in the Modeling Section of our Team Wiki. |
=== Goal 2 === | === Goal 2 === | ||
| - | The second goal of this project is to clone the ''Arabidopsis'' gene cinnamaldehyde-CoA reductase (gene 3) into an ''E. coli'' expression system and test for the production of cinnamaldehyde. Currently, we are building our construct using standard parts from the registry. The goal is to produce a newly engineered part containing an IPTG-inducible (Lac I) promoter, a standard RBS, | + | The second goal of this project is to clone the ''Arabidopsis'' gene cinnamaldehyde-CoA reductase (gene 3)into an ''E. coli'' expression system and test for the production of cinnamaldehyde. Currently, we are building our construct using standard parts from the registry. The goal is to produce a newly engineered part containing an IPTG-inducible (Lac I) promoter, a standard RBS, cinnamaldehyde-CoA reductase, and a standard double terminator. |
=== Experiment and Results for Goal 2=== | === Experiment and Results for Goal 2=== | ||
| Line 169: | Line 169: | ||
3. Conduct a three way ligation with intermediate parts from step 1 & 2 and a tetracycline resistant plasmid backbone (pSB3T5). Standard assembly 10. 3-WAY LIGATION HAS BEEN UNSUCCESSFUL TO DATE | 3. Conduct a three way ligation with intermediate parts from step 1 & 2 and a tetracycline resistant plasmid backbone (pSB3T5). Standard assembly 10. 3-WAY LIGATION HAS BEEN UNSUCCESSFUL TO DATE | ||
| - | 4. Alternative approach to step 3 is to perform a two-way ligation. Perform Restriciton Digest with Xba1 and Pst1 to isolate gene 3/double terminator, also run restriction digest with Spe1 and Pst1 Lac/RBS and clone | + | 4. Alternative approach to step 3 is to perform a two-way ligation. Perform Restriciton Digest with Xba1 and Pst1 to isolate gene 3/double terminator, also run restriction digest with Spe1 and Pst1 Lac/RBS and clone cinnamaldehyde-CoA reductase/double terminator into the chloromphenicol plasmid containing LacI+ RBS. IN PROGRESS |
=== Goal 3 === | === Goal 3 === | ||
| - | The third goal of this project is to clone the ''Arabidopsis'' gene 4-coumarate:CoA Ligase (gene 2) into an ''E. coli'' expression system and test for the production of cinnamoyl-CoA. Currently, we are building our | + | The third goal of this project is to clone the ''Arabidopsis'' gene 4-coumarate:CoA Ligase (gene 2) into an ''E. coli'' expression system and test for the production of cinnamoyl-CoA. Based on information obtained from our modeling studies, it is clear that the wild type plant 4-coumarate-CoA ligase has a very low affinity (i.e. high Km) for its substrate cinnamic acid. Fortunately we found an article published by Dr. Kombrink from the Max Planck Institute in Munchen, Germany that described the engineering of a 4-coumarate-CoA ligase with a forty fold decreased Km for cinnamic acid. Dr. Kombrink has generously provided us with the gene encoding this enzyme for use in this project. Therefore, we are currently working to express the wild type and engineered 4-coumarate-CoA ligase in ''E. coli.'' Currently, we are building our constructs using standard parts from the registry. The goal is to produce a newly engineered parts containing the Lac inducible (Lac I) promoter, a standard RBS, the wild type and engineered 4-coumarate-CoA ligase, and a standard double terminator. |
=== Experiment and Results for Goal 3=== | === Experiment and Results for Goal 3=== | ||
| - | 1. Conduct a three way ligation with 4-coumarate CoA Ligase | + | 1. Conduct a three way ligation with the engineered 4-coumarate:CoA Ligase, double terminator (BBa_B0014) and chloramphenicol resistant plasmid (pSB3C5). Follows standard assembly 10 protocol. DONE |
| - | 2. Conduct a three way ligation with BBa_K262000 and | + | 2. Conduct a three way ligation with BBa_K262000 and engineered 4-coumarate:CoA Ligase/double terminator and a tetracycline resistant plasmid backbone (pSB3T5). Standard assembly 10. 3-WAY LIGATION HAS BEEN UNSUCCESSFUL TO DATE DECIDED TO SKIP TO 2 WAY LIGATION APPROACH BELOW |
| - | + | 3. Alternative approach to step 2 is to perform a two-way ligation. Perform a restriciton digest of the engineered 4-coumarate:CoA Ligase/double terminator with Xba1 and Pst1, then isolate the engineered 4-coumarate:CoA Ligase/double terminator by gel extraction. Also run restriction digest with Spe1 and Pst1 to linearize BBa_K262000 and clone the engineered 4-coumarate:CoA Ligase/double terminator into BBa_K262000. IN PROGRESS | |
=== Goal 4 === | === Goal 4 === | ||
| - | + | The forth goal of this project is to construct an operon expressing all three genes encoding the complete enzyme complement of the cinnamaldehyde biosynthetic pathway in ''E. coli''. To achieve this we will clone the phenylalanine ammonia lyase gene (gene 1), the engineered 4-coumarate:CoA ligase gene (gene 2), and the cinnamoyl:CoA reductase gene (gene 3) into an ''E. coli'' expression system. | |
=== Experiment and Results for Goal 4=== | === Experiment and Results for Goal 4=== | ||
| - | 1. Conduct a three way ligation with 4-coumarate:CoA ligase (gene 2), RBS ( | + | 1. Conduct a three-way ligation with 4-coumarate:CoA ligase (gene 2), RBS (BBa_B0034) and chloramphenicol resistant plasmid (pSB3C5). Follows standard assembly 10 protocol. IN PROGRESS |
| - | 2. Conduct a three way ligation with BBa_K262000 and plasmid containing | + | 2. Conduct a three way ligation with BBa_K262000 and plasmid containing RBS/4-coumarate:CoA ligase. FAILED, REVISE STRATEGY |
=== Goal 5 === | === Goal 5 === | ||
| - | + | The fifth goal of this project is to develop ''in vitro'' enzyme assays to assess 4-coumarate-CoA ligase and cinnamoyl-CoA reductase activity in strains developed in Goals 2, 3, and 4. These assays will be important to verify the activity and level of expression of the engineered enzymes. Furthermore, the assays will be important to obtain kinetic information to apply to the model described in Goal 1. | |
| - | === | + | === Experiment and Results for Goal 5 === |
| - | + | ||
| - | SDS-PAGE showing 4-coumarate-CoA ligase protein expression (before it was made into a BioBrick part) under induction vs uninduced and wild type (W.T.) E. coli. An expression vector containing a gene for | + | 1) 4-Coumarate-CoA Ligase Assay: |
| + | The assay is designed to measure the production of cinnamoyl-CoA from cinnamic acid + ATP + CoA-SH by the enzyme 4-coumarate-CoA ligase. Cinnamoyl-CoA production is determined by the increase in absorbance at 311 nm, a signature absorbance maximun for the compound. | ||
| + | * Assay mixture: 3.3mg trans-cinnamic acid dissolved in 50uL ethanol, 2.0mg coenzyme A and 6.9mg adenosine triphosphate were suspended in 10mL 50mM Tris buffer 2.5mM magnesium chloride, pH 7.5. | ||
| + | * 50uL extract was added to 500uL assay mixture at room temperature in a quartz cuvette, and change in absorbance was measured at 311nm. | ||
| + | |||
| + | |||
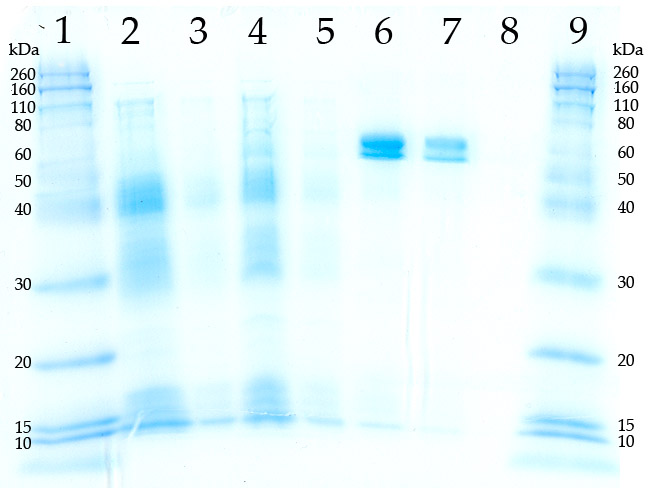
| + | <div style="text-align:center">[[Image:IGEM_Gene_2_Gel.jpg]]</div> | ||
| + | |||
| + | |||
| + | Coomassie stained SDS-PAGE gel showing 4-coumarate-CoA ligase protein expression (before it was made into a BioBrick part) under induction vs uninduced and wild type (W.T.) E. coli. An expression vector, PL N256A (donated by Dr. Kombrink at the Max Planck Institute, Munchen, Germany), containing a modified gene for 4-coumarate:CoA ligase, an IPTG inducible promoter and a 6xHis tag was used to transform E. coli, BL21 strain cells. Tagged protein in lanes 6 and 7 were purified using a Ni column ("HiTrap nickel chelating column," GE Biosciences). Lanes are as follows: | ||
# Molecular weight marker | # Molecular weight marker | ||
| Line 209: | Line 217: | ||
# Molecular weight marker | # Molecular weight marker | ||
| - | Our data shows that wild type E. coli display no 4-coumarate:CoA ligase expression. Lanes 4, 5, 6 and 7 show that the transformants do express 4-coumarate-CoA ligase. | + | Our data shows that wild type E. coli display no 4-coumarate:CoA ligase expression. Lanes 4, 5, 6 and 7 show that the transformants do express the 66 kD 4-coumarate-CoA ligase protein. |
| - | [[Image:IGEM2.JPG]] | + | <div style="text-align:center">[[Image:IGEM2.JPG]]</div> |
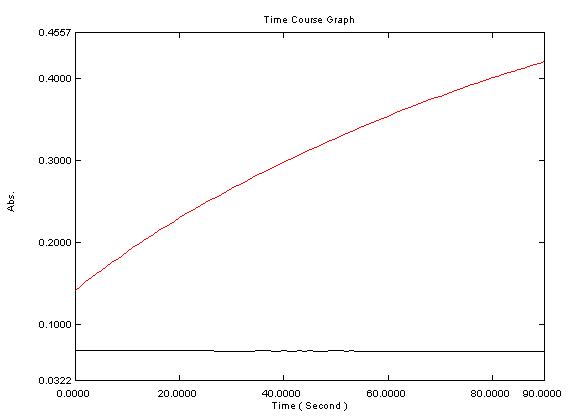
| - | Time course of 4-coumarate:CoA ligase activty. 50uL of clarified E. coli extract was added to 500uL of enzyme assay solution. Black line = W.T. E. coli. Red line = E. coli transformed with | + | Time course of 4-coumarate:CoA ligase activty. 50uL of clarified E. coli extract was added to 500uL of enzyme assay solution. Black line = W.T. E. coli. Red line = E. coli transformed with PL N256A. |
| - | [[Image:Igem3.JPG]] | + | <div style="text-align:center">[[Image:Igem3.JPG]]</div> |
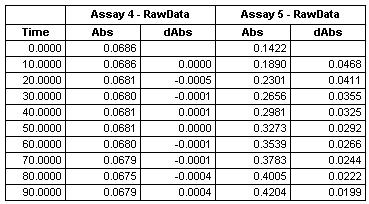
Absorbance change/10 s interval given in table: assay 4 = wild type. Assay 5 = E. coli transformed with PL N256A. | Absorbance change/10 s interval given in table: assay 4 = wild type. Assay 5 = E. coli transformed with PL N256A. | ||
| + | |||
| + | 2) Cinnamoyl-CoA Reductase Assay: IN PROGRESS | ||
=== Goal 6 === | === Goal 6 === | ||
| - | + | The sixth goal of this project is to transform one or more genes of the cinnamaldehyde pathway created in goals 2, 3, and 4 into the aquatic plant species ''Wolffia'' and ''Lemna minor''. We hope to enhance the endogenous cinnamaldehyde pathway in these plants (which normally make very little cinnamaldehyde) in order to produce more of our desired product. Ultimately, the products produced by these plants could possibly be used as an adult mosquito repellent and larvicide to certain mosquito larvae. | |
=== Experiment and Results for Goal 6=== | === Experiment and Results for Goal 6=== | ||
| - | 1. Conduct TOPO Cloning Reaction to put | + | 1. Conduct TOPO Cloning Reaction to put cinnamoyl:CoA reductase into pEntrD TOPO Vector (Shuttle vector which will allow us to put cinnamoyl:CoA reductase into a binary plant vector) DONE |
| - | 2. Conduct LR Clonase Reaction to transfer | + | 2. Conduct LR Clonase Reaction to transfer cinnamoyl:CoA reductase from pEntrD Vector to pK7WG2D.1 Binary Vector. DONE |
3. Use Binary Vector pK7WG2D.1 to grow Agrobacterium EHA105 strain. Use this strain to infect Wolffia. IN PROGRESS | 3. Use Binary Vector pK7WG2D.1 to grow Agrobacterium EHA105 strain. Use this strain to infect Wolffia. IN PROGRESS | ||
| Line 246: | Line 256: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | |||
| - | |||
==References== | ==References== | ||
| Line 271: | Line 279: | ||
#Zhu, J., Zeng, X., Yanma, Liu, T., Qian, K., Han, Y., Xue, S., Tucker, B., Schultz, G., Coats, J., Rowley, W., Zhang, A. 2006. Adult repellency and larvicidal activity of five plant essential oils against mosquitoes. Jounral of the American Mosquito Control Association. 22, 515-522. | #Zhu, J., Zeng, X., Yanma, Liu, T., Qian, K., Han, Y., Xue, S., Tucker, B., Schultz, G., Coats, J., Rowley, W., Zhang, A. 2006. Adult repellency and larvicidal activity of five plant essential oils against mosquitoes. Jounral of the American Mosquito Control Association. 22, 515-522. | ||
#Zhu, J., Zeng, X., O'Neal, M., Schultz, G., Tucker, B., Coats, J., Bartholomay, L., Xue, R.D. 2008. Mosquito Larvicidal activity of botanical-based mosquito repellents. Journal of the American Mosquito Control Association. 24, 161-168. | #Zhu, J., Zeng, X., O'Neal, M., Schultz, G., Tucker, B., Coats, J., Bartholomay, L., Xue, R.D. 2008. Mosquito Larvicidal activity of botanical-based mosquito repellents. Journal of the American Mosquito Control Association. 24, 161-168. | ||
| + | ==Special Acknowledgements== | ||
| + | We would like to thank Dr. Kombrink from the Max Planck Institute in Munchen, Germany for his generous gift of the engineered 4-coumarate-CoA ligase gene. We also thank Dr. Todd Michaels from the Waksmann Institute at Rutgers University for providing our team with duckweed samples and protocols. We would also like to thank the University of Nevada College of Agriculture, Biotechnology and Natural Resources, the University of Nevada Department of Biochemistry and Molecular Biology, the University of Nevada Vice President of Research Marsh Read, Nevada INBRE, and the Associated Students of the University of Nevada for their financial support of the 2009 Nevada iGEM Team. | ||
| + | |||
| + | </div> | ||
| + | |||
<html> | <html> | ||
<div> | <div> | ||
<img src="https://static.igem.org/mediawiki/2009/6/63/Bottom.jpg" border="0" alt="" width="965" /> | <img src="https://static.igem.org/mediawiki/2009/6/63/Bottom.jpg" border="0" alt="" width="965" /> | ||
</div> | </div> | ||
| - | |||
| - | |||
| - | |||
</html> | </html> | ||
Latest revision as of 03:50, 22 October 2009
Overall project
Cinnamicide: Producing a Natural Insecticide against Mosquito Larvae in E. coli and Duckweed
Cinnamaldehyde is a natural insecticide against mosquito larvae that shows low toxicity towards other organisms. The objective of this project is to engineer the cinnamaldehyde biosynthetic pathway into E. coli to develop an inexpensive and readily available source of this compound. By introducing the genes encoding phenylalanine-ammonia lyase (gene 1), 4-coumarate:CoA ligase (gene 2), and cinnamoyl-CoA reductase (gene 3), it should be possible to produce cinnamaldehyde from available phenylalanine in E. coli. Once we have demonstrated that we can produce cinnamaldehyde in E. coli, we will engineer cinnamaldehyde production in duckweed (Wolffia or Lemna species), a small aquatic plant. Because mosquito larvae feed on duckweed detritus, the engineered plant will serve as an excellent vehicle to deliver cinnamaldehyde for mosquito control.
Safety Questionnaire
Would any of your project ideas raise safety issues in terms of:
- researcher safety,
- public safety, or
- environmental safety?
At this point in the project, it will not raise any safety concerns in terms of researcher safety, public safety, or environmental safety. Future portions of this project pertaining to transformation of E. coli and duckweed could raise environmental issues and proper containment monitoring will need to be addressed.
Is there a local biosafety group, committee, or review board at your institution?
Yes, there is an Institutional Biosafety Committee (IBC) at our institution which is part of the Environmental Health & Safety (EH&S) program. We also collaborated with Scott Munsen of the Washoe County Vector Control Program.
What does your local biosafety group think about your project?
Our local biosafety group approved our project design and subsequent facilitation of research. Scott Munsen of the Washoe County Vector Control Program believes this could be a feasible pest control and will aid in the larvicidal tests of cinnamaldehyde against various mosquito species to verify its larvicidal activity and to ensure we are not harming non-target organisms.
Do any of the new BioBrick parts that you made this year raise any safety issues?
No, none of our new BioBrick parts that we made this year raise any safety issues. The genes we are using are naturally occurring genes expressed in plants and other organisms that do not pose any risk.
Project Details
Why Produce an Insecticide against Mosquitoes?
It is estimated that every year over 350-500 million people are infected by diseases spread by mosquitoes, and over 1 million of them die as a result. Current insecticides being used which include dichlorodiphenyltrichloroethane (DDT), organophosphates, and others, cause environmental damage to non-target organisms, including humans. Many mosquito species gain resistance to these insecticides, so new ways to combat mosquitoes that transmit human diseases need to be produced. In order to replace these general synthetic insecticides, our proposed project is to produce a natural, low toxicity mosquito larvicide through the production of cinnamaldehyde. Finding a cost-effective, environmentally friendly manner to manage mosquito populations is a key aim of this project.
Goal 1
Develop a mathematical model to predict pathway flux based on enzyme kinetic values and levels of gene expression. The information obtained from this model will help rationalize engineering design choices to optimize cinnamaldehyde production in E.coli.
Experiment and Results for Goal 1
1. Using Mathematica, a model was developed to describe Michaelis-Menten steady-state kinetics for the engineered cinnamaldehyde biosynthetic pathway. The published Km and Vmax values for the plant enzyme, phenylalanine-ammonia lyase, 4-coumarate:CoA ligase and cinnamoyl-CoA reductase (feruloyl-CoA reductase), were used to build our model. Using wild type kinetic parameters and the assumption that all three enzymes were present in equal amounts, the model predicted that a biosynthetic bottleneck will occur at 4-coumarate:CoA ligase. This problem appears to be primarily due to the high Km value of the enzyme for its substrate cinnamic acid. Therefore, we used the model to test two possible means of increasing pathway flux: 1) increasing 4-coumarate:CoA ligase levels and 2) decreasing 4-coumarate:CoA ligase’s Km for cinnamic acid. The results produced from the model showed that by increasing the 4-coumarate:CoA ligase concentration ten-fold relative to the other enzymes in the pathway, or by decreasing the Km forty-fold, the barrier could be bypassed in this way. the results of these studies are shown in the Modeling Section of our Team Wiki.
Goal 2
The second goal of this project is to clone the Arabidopsis gene cinnamaldehyde-CoA reductase (gene 3)into an E. coli expression system and test for the production of cinnamaldehyde. Currently, we are building our construct using standard parts from the registry. The goal is to produce a newly engineered part containing an IPTG-inducible (Lac I) promoter, a standard RBS, cinnamaldehyde-CoA reductase, and a standard double terminator.
Experiment and Results for Goal 2
1. Conduct a three way ligation with LacI promoter (BBa_R0011), RBS (part BBa_B0034) and a chloramphenicol resistant plasmid backbone (pSB3C5) using the standard assembly 10 protocol. DONE created intermediate part BBa_K262000
2. Conduct a three way ligation with cinnamoyl-CoA reductase (gene 3), double terminator (BBa_B0014) and chloramphenicol resistant plasmid (pSB3C5). Follows standard assembly 10 protocol. DONE created intermediate part
BBa_K262002
3. Conduct a three way ligation with intermediate parts from step 1 & 2 and a tetracycline resistant plasmid backbone (pSB3T5). Standard assembly 10. 3-WAY LIGATION HAS BEEN UNSUCCESSFUL TO DATE
4. Alternative approach to step 3 is to perform a two-way ligation. Perform Restriciton Digest with Xba1 and Pst1 to isolate gene 3/double terminator, also run restriction digest with Spe1 and Pst1 Lac/RBS and clone cinnamaldehyde-CoA reductase/double terminator into the chloromphenicol plasmid containing LacI+ RBS. IN PROGRESS
Goal 3
The third goal of this project is to clone the Arabidopsis gene 4-coumarate:CoA Ligase (gene 2) into an E. coli expression system and test for the production of cinnamoyl-CoA. Based on information obtained from our modeling studies, it is clear that the wild type plant 4-coumarate-CoA ligase has a very low affinity (i.e. high Km) for its substrate cinnamic acid. Fortunately we found an article published by Dr. Kombrink from the Max Planck Institute in Munchen, Germany that described the engineering of a 4-coumarate-CoA ligase with a forty fold decreased Km for cinnamic acid. Dr. Kombrink has generously provided us with the gene encoding this enzyme for use in this project. Therefore, we are currently working to express the wild type and engineered 4-coumarate-CoA ligase in E. coli. Currently, we are building our constructs using standard parts from the registry. The goal is to produce a newly engineered parts containing the Lac inducible (Lac I) promoter, a standard RBS, the wild type and engineered 4-coumarate-CoA ligase, and a standard double terminator.
Experiment and Results for Goal 3
1. Conduct a three way ligation with the engineered 4-coumarate:CoA Ligase, double terminator (BBa_B0014) and chloramphenicol resistant plasmid (pSB3C5). Follows standard assembly 10 protocol. DONE
2. Conduct a three way ligation with BBa_K262000 and engineered 4-coumarate:CoA Ligase/double terminator and a tetracycline resistant plasmid backbone (pSB3T5). Standard assembly 10. 3-WAY LIGATION HAS BEEN UNSUCCESSFUL TO DATE DECIDED TO SKIP TO 2 WAY LIGATION APPROACH BELOW
3. Alternative approach to step 2 is to perform a two-way ligation. Perform a restriciton digest of the engineered 4-coumarate:CoA Ligase/double terminator with Xba1 and Pst1, then isolate the engineered 4-coumarate:CoA Ligase/double terminator by gel extraction. Also run restriction digest with Spe1 and Pst1 to linearize BBa_K262000 and clone the engineered 4-coumarate:CoA Ligase/double terminator into BBa_K262000. IN PROGRESS
Goal 4
The forth goal of this project is to construct an operon expressing all three genes encoding the complete enzyme complement of the cinnamaldehyde biosynthetic pathway in E. coli. To achieve this we will clone the phenylalanine ammonia lyase gene (gene 1), the engineered 4-coumarate:CoA ligase gene (gene 2), and the cinnamoyl:CoA reductase gene (gene 3) into an E. coli expression system.
Experiment and Results for Goal 4
1. Conduct a three-way ligation with 4-coumarate:CoA ligase (gene 2), RBS (BBa_B0034) and chloramphenicol resistant plasmid (pSB3C5). Follows standard assembly 10 protocol. IN PROGRESS
2. Conduct a three way ligation with BBa_K262000 and plasmid containing RBS/4-coumarate:CoA ligase. FAILED, REVISE STRATEGY
Goal 5
The fifth goal of this project is to develop in vitro enzyme assays to assess 4-coumarate-CoA ligase and cinnamoyl-CoA reductase activity in strains developed in Goals 2, 3, and 4. These assays will be important to verify the activity and level of expression of the engineered enzymes. Furthermore, the assays will be important to obtain kinetic information to apply to the model described in Goal 1.
Experiment and Results for Goal 5
1) 4-Coumarate-CoA Ligase Assay: The assay is designed to measure the production of cinnamoyl-CoA from cinnamic acid + ATP + CoA-SH by the enzyme 4-coumarate-CoA ligase. Cinnamoyl-CoA production is determined by the increase in absorbance at 311 nm, a signature absorbance maximun for the compound.
- Assay mixture: 3.3mg trans-cinnamic acid dissolved in 50uL ethanol, 2.0mg coenzyme A and 6.9mg adenosine triphosphate were suspended in 10mL 50mM Tris buffer 2.5mM magnesium chloride, pH 7.5.
- 50uL extract was added to 500uL assay mixture at room temperature in a quartz cuvette, and change in absorbance was measured at 311nm.
Coomassie stained SDS-PAGE gel showing 4-coumarate-CoA ligase protein expression (before it was made into a BioBrick part) under induction vs uninduced and wild type (W.T.) E. coli. An expression vector, PL N256A (donated by Dr. Kombrink at the Max Planck Institute, Munchen, Germany), containing a modified gene for 4-coumarate:CoA ligase, an IPTG inducible promoter and a 6xHis tag was used to transform E. coli, BL21 strain cells. Tagged protein in lanes 6 and 7 were purified using a Ni column ("HiTrap nickel chelating column," GE Biosciences). Lanes are as follows:
- Molecular weight marker
- W.T. E. coli, crude extract, 50ug protein
- W.T. E. coli, crude extract, 10ug protein
- Transformed E. Coli, uniduced crude protein extract, 50ug protein
- Transformed E. Coli, uniduced crude protein extract, 10ug protein
- IPTG induced transformed E. coli, Ni Column purified, Coumarate:CoA ligase extract, 50ug protein
- IPTG induced transformed E. coli, Ni Column purified, Coumarate:CoA ligase extract, 10ug protein
- Blank
- Molecular weight marker
Our data shows that wild type E. coli display no 4-coumarate:CoA ligase expression. Lanes 4, 5, 6 and 7 show that the transformants do express the 66 kD 4-coumarate-CoA ligase protein.
Time course of 4-coumarate:CoA ligase activty. 50uL of clarified E. coli extract was added to 500uL of enzyme assay solution. Black line = W.T. E. coli. Red line = E. coli transformed with PL N256A.
Absorbance change/10 s interval given in table: assay 4 = wild type. Assay 5 = E. coli transformed with PL N256A.
2) Cinnamoyl-CoA Reductase Assay: IN PROGRESS
Goal 6
The sixth goal of this project is to transform one or more genes of the cinnamaldehyde pathway created in goals 2, 3, and 4 into the aquatic plant species Wolffia and Lemna minor. We hope to enhance the endogenous cinnamaldehyde pathway in these plants (which normally make very little cinnamaldehyde) in order to produce more of our desired product. Ultimately, the products produced by these plants could possibly be used as an adult mosquito repellent and larvicide to certain mosquito larvae.
Experiment and Results for Goal 6
1. Conduct TOPO Cloning Reaction to put cinnamoyl:CoA reductase into pEntrD TOPO Vector (Shuttle vector which will allow us to put cinnamoyl:CoA reductase into a binary plant vector) DONE
2. Conduct LR Clonase Reaction to transfer cinnamoyl:CoA reductase from pEntrD Vector to pK7WG2D.1 Binary Vector. DONE
3. Use Binary Vector pK7WG2D.1 to grow Agrobacterium EHA105 strain. Use this strain to infect Wolffia. IN PROGRESS
References
- Arriaga, A.M.C., Rodrigues, F.E.A., Lemos, T.L.G., de Oliveria, M.C.F., Lima, J.Q., Santiago, G.M.P., Braz-Filho, R., Mafezoli, J. 2007.Composition and larvicidal activity of essential oils from Stemodia maritima L. NPC. 2, 1237-1239.
- Beuerle, T., Pichersky, E. 2002. Enzymatic synthesis and purification of aromatic coenzyme A esters. Analytical Biochemistry. 302, 305-312.
- bin Jantan, I., Yalvema, M.F., Ahmad, N.W., Jamal, J.A. 2005. Insecticidal activities of the leaf oils of eight Cinnamomum species against Aedes aegypti and Aedes albopictus. Pharmaceutical Biology. 43, 526-532.
- Chang, K.S., Tak, J.H., Kim, S.I., Lee, W.J., Ahn, Y.J. 2006. Repellency of Cinnamomum cassia bark compounds and cream containing casia oil to Aedes aegypti (Diptera: Culicidae) under laboratory and indoor conditions. 62, 1032-1038.
- Cheng, S.S., Liu, J.Y., Tsai, K.H., Chen, W.J., Chang, S.T. 2004. Chemical composition and mosquito larvicidal activity of essential oils from leaves of different Cinnamomum osmophloeum provenances. J. Agric. Food Chem. 52, 4395-4400.
- Cheng, S., Liu, J.Y., Huang, C.G., Hsui, Y.R., Chen, W.J, Chang, S.T. 2008. Insecticidal activities of leaf essential oils from Cinnamomum osmophloeum against three mosquito species. Bioresource Technology. 100, 457-464.
- Hamberger, B., Hahlbrock, K. 2003. The 4-coumarate:CoA ligase gene family in Arabidopsis thaliana comprises one rare, sinapate-activating and three commonly occurring isoenzymes. PNAS. 101, 2209-2214.
- Kim, N.J., Byun, S.G., Cho, J.E., Chung, K, Ahn, Y.J. 2008. Larvicidal activity of Kaempferia galanga rhizome phenylpropanoids towards three mosquito species. Pest Manag Sci. 64, 857-862.
- Lee, E.J., Kim, J.R., Choi, D.R, Ahn, Y.J. 2008. Toxicity of Cassia and Cinnamomum Oil compounds and Cinnamaldehyde-related compounds to Sitophilus oryzae (Coleoptera: curculionidae). Entomological Society of America. 101, 1960-1966.
- Li, J., Jain, M., Vunsh, R., Vishnevetsky, J., Hanania, U., Flaishman, M., Perl, A., Edelman, M. 2004. Callus induction and regeneration in Spirodela and Lemna. Plant Cell Rep. 22, 457-464.
- Mulquiney, P.J., Kuchel, P.W. Modeling Metabolism with Mathematica. CRC Press: 2003.
- Rajkumar, S., Jebanesan, A. 2009. Larvicidal and oviposition activity of Cassia obtusifolia Linn (Family: Leguminosae) leaf extract against malarial vector, Anopheles stephensi Liston (Diptera: Culicidae). Parasitol Res. 104, 337-340.
- Santos, H.S. Santiago, G.M.P., de Oliveria, J.P.P., Arriaga, A.M.C., Marques, DD., Lemos, T.L.G. 2007. NPC. 2, 1233-1236.
- Schneider, K., Hovel, K., Witzel, K., Hamberger, B., Schombur, D., Kombrink, E., Stuible, H.P. 2003. The substrate specificity-determining amino acid code of 4-coumarate:CoA ligase. PNAS. 100, 8601-8606.
- Stuible, H.P., Buttner, D., Ehlting, J., Hahlbrock, K., Kombrink, E. 2000. Mutational analysis of 4-coumarate:CoA ligase identifies functionally important amino acids and verifies its close relationship to other adenylate-forming enzymes. FEBS Letters. 467, 117-122.
- Williams, L.A.D., Porter, R.B., Junor, G.O. 2007. Biological activities of selected essential oils. NPC. 2, 1295-1296.
- Yamamoto, Y.T., Rajbhandari, N., Lin, X., Bergmann, B.A., Nishimura, Y., Stomp, A.M. 2000. Genetic transformation of duckweed Lemna Gibba and Lemna Minor. In Vitro Cell. Dev. Biol. 37. 349-353.
- Yun, M.S., Chen, W., Deng, F., Yogo, Y. 2005. Differential properties of 4-coumarate : CoA ligase related to growth suppression of chalcone in maize and rice. Plant Growth Regulation. 46, 169-176.
- Zhu, J., Zeng, X., Yanma, Liu, T., Qian, K., Han, Y., Xue, S., Tucker, B., Schultz, G., Coats, J., Rowley, W., Zhang, A. 2006. Adult repellency and larvicidal activity of five plant essential oils against mosquitoes. Jounral of the American Mosquito Control Association. 22, 515-522.
- Zhu, J., Zeng, X., O'Neal, M., Schultz, G., Tucker, B., Coats, J., Bartholomay, L., Xue, R.D. 2008. Mosquito Larvicidal activity of botanical-based mosquito repellents. Journal of the American Mosquito Control Association. 24, 161-168.
Special Acknowledgements
We would like to thank Dr. Kombrink from the Max Planck Institute in Munchen, Germany for his generous gift of the engineered 4-coumarate-CoA ligase gene. We also thank Dr. Todd Michaels from the Waksmann Institute at Rutgers University for providing our team with duckweed samples and protocols. We would also like to thank the University of Nevada College of Agriculture, Biotechnology and Natural Resources, the University of Nevada Department of Biochemistry and Molecular Biology, the University of Nevada Vice President of Research Marsh Read, Nevada INBRE, and the Associated Students of the University of Nevada for their financial support of the 2009 Nevada iGEM Team.

 "
"