EPF-Lausanne/31 August 2009
From 2009.igem.org
(→Wet Lab) |
(→Wet Lab) |
||
| Line 42: | Line 42: | ||
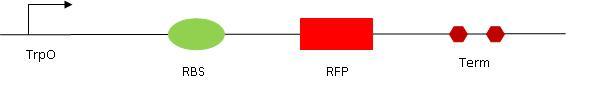
[[Image:RO1.jpg|center|Read out 1 design]] | [[Image:RO1.jpg|center|Read out 1 design]] | ||
| + | '''Digestion''' of the concentrated Klenow PCR. After digestion, both gel extraction and purification kit will be tried. | ||
| - | ===RO2 | + | We did a '''gel''' to check the digestion assay of TrpO and the samples from Saturday. TrpO, BB5, RO2#8 and the double transformation seems okay. The double constructs didn't work, for both clones (it seems not to have included the BB, all patterns are looking the same as the positive RO2 control). |
| + | |||
| + | |||
| + | '''Transformation''' of the following plasmids from kit plate n°1 have been transformed in DH5a : I13602 (on pSB1A2) and J04450 (on pSB1C3). The first plasmid is to check if DH5a has endogenous TetR gene (if it doesn't - as supposed - then the bacterium will express CFP, no matter if we had ATC or not. ATC needs to have the TetR protein to bind to and have an effect). The second plasmid is to have a test both on IPTG induction (on LacI as in our construction) and also to have a positive control for RFP. | ||
| + | |||
| + | The host vector (I13507) containing RFP has been digested with E and X. After incubation, purification using the purelink kit. | ||
| + | |||
| + | '''Gel extraction''' of the TrpO digestion products, using the Giagen gel extraction kit protocol. | ||
| + | |||
| + | '''Ligation''' of RO1 overnight. | ||
| + | |||
| + | ===RO2 genetic circuit=== | ||
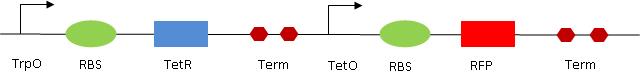
[[Image:RO2.jpg|center|Read out 2 design]] | [[Image:RO2.jpg|center|Read out 2 design]] | ||
| - | ===LovTAP | + | ===LovTAP genetic circuit=== |
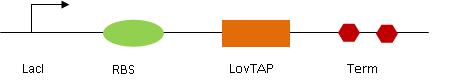
[[Image:LovTAP.jpg|center|LovTAP design]] | [[Image:LovTAP.jpg|center|LovTAP design]] | ||
Latest revision as of 12:03, 8 September 2009
Contents |
Wet Lab
Digestion
The Klenow fragments that have been amplified by PCR (after the Klenow reaction) will be digested with SpeI. We have to check if it is indeed the TrpO (presence of the two S sites).
Results of the overweekend cultures
The idea was to find a new medium without any TRY on it, but on which the cells can still grow.
The minimal media + arginine didn't show any result (nothing grew).
The tubes induced by Try didn't change colour over the week-end. The two that were already red are still red and the others didn't change colour.
RO1 construction
Digestion of the concentrated Klenow PCR. After digestion, both gel extraction and purification kit will be tried.
We did a gel to check the digestion assay of TrpO and the samples from Saturday. TrpO, BB5, RO2#8 and the double transformation seems okay. The double constructs didn't work, for both clones (it seems not to have included the BB, all patterns are looking the same as the positive RO2 control).
Transformation of the following plasmids from kit plate n°1 have been transformed in DH5a : I13602 (on pSB1A2) and J04450 (on pSB1C3). The first plasmid is to check if DH5a has endogenous TetR gene (if it doesn't - as supposed - then the bacterium will express CFP, no matter if we had ATC or not. ATC needs to have the TetR protein to bind to and have an effect). The second plasmid is to have a test both on IPTG induction (on LacI as in our construction) and also to have a positive control for RFP.
The host vector (I13507) containing RFP has been digested with E and X. After incubation, purification using the purelink kit.
Gel extraction of the TrpO digestion products, using the Giagen gel extraction kit protocol.
Ligation of RO1 overnight.
RO2 genetic circuit
LovTAP genetic circuit
People in the lab
Basile


 "
"