EPF-Lausanne/11 October 2009
From 2009.igem.org
(New page: {{EPF-Lausanne09}} <div CLASS="epfltrick">__TOC__ </div><div CLASS="epfl09"> <html> <body> <form action="input_button.htm"> <p align="right"> <input type="button" name="lien" value="10 Oc...) |
(→Wet Lab) |
||
| (6 intermediate revisions not shown) | |||
| Line 27: | Line 27: | ||
==Wet Lab== | ==Wet Lab== | ||
| + | '''Tests on RO1.1+BB1 DH5 alpha double-transformants? which until now didn't work:''' | ||
| - | |||
| + | Reinoculated 0.5mL of the overnight culture into 3.5mL of fresh LB. We did 4 different conditions: | ||
| + | |||
| + | - +light +IPTG -Trp | ||
| + | |||
| + | - -light +IPTG -Trp | ||
| + | |||
| + | - -light -IPTG +Trp | ||
| + | |||
| + | - -light -IPTG -Trp | ||
| + | |||
| + | Left cells to incubate in respective experimental conditions for about 1h30, then measured OD and fluorescence with the plate reader. The obtained results are the following: no fluorescence at all from the cells (they had the same values as for LB alone), so apparently either the double-transformants don't work, or the cells are not double-transformants, which we'll check with the digestion assay and gel (see below). | ||
| + | |||
| + | |||
| + | |||
| + | To make yet another test, we took what remained of the RO1.1+BB1 Dh5 alpha overnight liquid culture and miniprepped it. We then did a digestion assay by cutting with enzymes SpeI (which should cut in the suffixes and in the Trp promoter) and PstI (cuts in the suffix and in the LovTap). We then ran the digestion products on a gel to check the lengths of the fragments. From this we could conclude that in reality we don't have double-transformants. So we'll have to redo transformations and new experiments with other cell strains. | ||
| + | |||
| + | |||
| + | |||
| + | '''TrpR-mutated strains''' | ||
| + | |||
| + | |||
| + | |||
| + | Did a colony PCR on 6 clones each from the RO2.4+BB1 and RO1.1+BB1 JRG 1046 strain to look for double-transformants. Ran a gel. We saw that we might have double transformants, but since some bands are very close to each other (RO1=1000 bp and LovTap=1100bp) we can't be 100% sure. So we made cultures in LB of 3/6 clones for every read-out system, which we'll miniprep tomorrow and use to do a digestion assay. | ||
| + | |||
| + | For the same double-transformants but in the JRG 465 strain, we had bacterial lawns, so we did dilutions of these on new plates, and left them to incubate at 37°C. | ||
| + | |||
| + | |||
| + | |||
| + | '''RO2.4+BB1 testing''' | ||
| + | |||
| + | |||
| + | |||
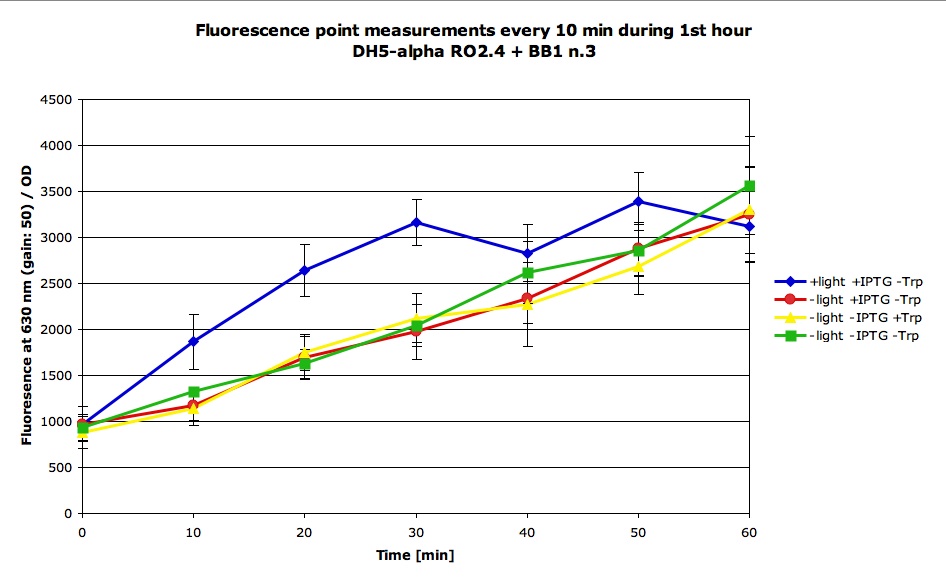
| + | Reinoculated 0.5 mL of the overnight culture of RO2.4+BB1 DH5 alpha strain into 3.5 mL of fresh LB, and put the cells in 4 conditions: | ||
| + | |||
| + | - +light +IPTG -Trp | ||
| + | |||
| + | - -light +IPTG -Trp | ||
| + | |||
| + | - -light -IPTG +Trp | ||
| + | |||
| + | - -light -IPTG -Trp | ||
| + | |||
| + | We prepared duplicates of all conditions and did the following experiments: | ||
| + | |||
| + | - Took measures of the OD and fluorescence every 10 min starting at 0 min exposure to conditions and going up to 60 min. The results are the following: | ||
| + | |||
| + | [[Image:Snapshot 2009-10-12 22-49-45.jpg|center|thumb|upright=4|RO2 semi-dynamic]] | ||
| + | |||
| + | - Left the cells to incubate in conditions for 2h and took a point measurement of OD and flurorescence to check the reproductibility of our past results. We obtain the following graph: coming soon... | ||
| + | |||
| + | - After cells had incubated for about 2h and had a maximal expression of RFP, left cells in the plate reader for a kinetic measurement of fluorescence, to measure the degradation of RFP. | ||
| + | |||
| + | ==People in the lab== | ||
| + | |||
| + | Tu, Gab, Caroline | ||
<html><center><a href="https://2009.igem.org/EPF-Lausanne/10_October_2009"><img src="https://static.igem.org/mediawiki/2009/thumb/6/61/Flèche_gauche.png/70px-Flèche_gauche.png"></a> | <html><center><a href="https://2009.igem.org/EPF-Lausanne/10_October_2009"><img src="https://static.igem.org/mediawiki/2009/thumb/6/61/Flèche_gauche.png/70px-Flèche_gauche.png"></a> | ||
Latest revision as of 20:58, 12 October 2009
Contents |
Wet Lab
Tests on RO1.1+BB1 DH5 alpha double-transformants? which until now didn't work:
Reinoculated 0.5mL of the overnight culture into 3.5mL of fresh LB. We did 4 different conditions:
- +light +IPTG -Trp
- -light +IPTG -Trp
- -light -IPTG +Trp
- -light -IPTG -Trp
Left cells to incubate in respective experimental conditions for about 1h30, then measured OD and fluorescence with the plate reader. The obtained results are the following: no fluorescence at all from the cells (they had the same values as for LB alone), so apparently either the double-transformants don't work, or the cells are not double-transformants, which we'll check with the digestion assay and gel (see below).
To make yet another test, we took what remained of the RO1.1+BB1 Dh5 alpha overnight liquid culture and miniprepped it. We then did a digestion assay by cutting with enzymes SpeI (which should cut in the suffixes and in the Trp promoter) and PstI (cuts in the suffix and in the LovTap). We then ran the digestion products on a gel to check the lengths of the fragments. From this we could conclude that in reality we don't have double-transformants. So we'll have to redo transformations and new experiments with other cell strains.
TrpR-mutated strains
Did a colony PCR on 6 clones each from the RO2.4+BB1 and RO1.1+BB1 JRG 1046 strain to look for double-transformants. Ran a gel. We saw that we might have double transformants, but since some bands are very close to each other (RO1=1000 bp and LovTap=1100bp) we can't be 100% sure. So we made cultures in LB of 3/6 clones for every read-out system, which we'll miniprep tomorrow and use to do a digestion assay.
For the same double-transformants but in the JRG 465 strain, we had bacterial lawns, so we did dilutions of these on new plates, and left them to incubate at 37°C.
RO2.4+BB1 testing
Reinoculated 0.5 mL of the overnight culture of RO2.4+BB1 DH5 alpha strain into 3.5 mL of fresh LB, and put the cells in 4 conditions:
- +light +IPTG -Trp
- -light +IPTG -Trp
- -light -IPTG +Trp
- -light -IPTG -Trp
We prepared duplicates of all conditions and did the following experiments:
- Took measures of the OD and fluorescence every 10 min starting at 0 min exposure to conditions and going up to 60 min. The results are the following:
- Left the cells to incubate in conditions for 2h and took a point measurement of OD and flurorescence to check the reproductibility of our past results. We obtain the following graph: coming soon...
- After cells had incubated for about 2h and had a maximal expression of RFP, left cells in the plate reader for a kinetic measurement of fluorescence, to measure the degradation of RFP.
People in the lab
Tu, Gab, Caroline


 "
"