Team:MoWestern Davidson/project wetlab
From 2009.igem.org
| Line 27: | Line 27: | ||
[[Image:RFPFluorescence.png|none|750px|]] | [[Image:RFPFluorescence.png|none|750px|]] | ||
| - | [[Image:RFPpellets.jpg]] | + | [[Image:RFPpellets.jpg|none|750px|]] |
Revision as of 19:55, 27 July 2009

We began our project design by constructing 12 suppressor tRNAs by oligo assembly. Suppressor tRNA genes were ligated with the pBad promoter to allow weak constitutive and inducible tRNA expression.
Choosing A Promoter
One important consideration in our project design was choosing the promoter(s) that would induce a notable phenotype. For controlling our suppressor tRNA expression, we wanted a promoter that would transcribe enough tRNA to cause a high probability of suppression but not so much tRNA that cells would become sick. For controlling our reporters with a leading logical clause, we wanted a promoter that would have strong enough transcription to allow more chances for suppression, and thereby more reporter expression.
We decided to use a weak-medium promoter, pBad, to express our suppressor tRNAs. This promoter choice allow us to induce greater expression with L-arabinose. We found that suppressor tRNAs controlled by pBad is not lethal to E. coli and allows for normal culture growth.
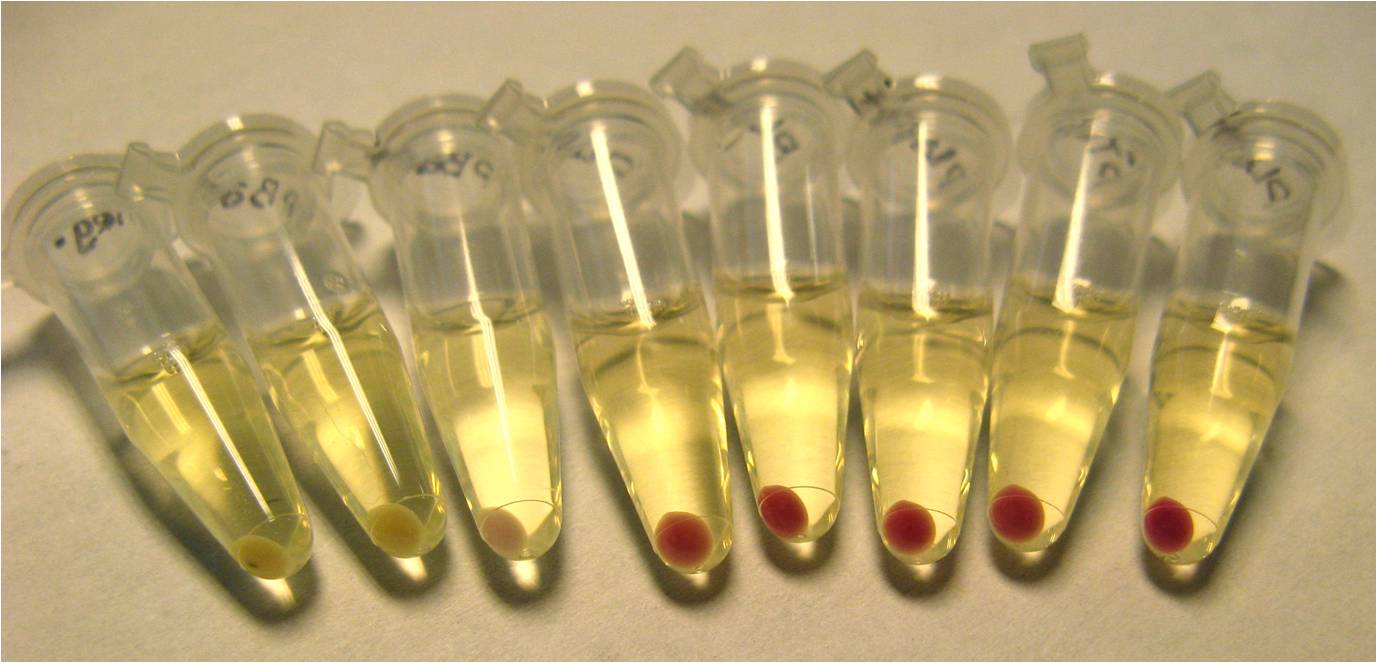
In order to have a visible expression of RFP, we constructed parts with RBS-RFP under the control of 4 different commonly-used promoters:
- pBad (Potential induction with L-arabinose)
- pLacI (also called pLac. Potential induction with IPTG)
- pLacIQ (Potential induction with IPTG)
- pTet
We found that pLac induced with IPTG caused the greatest expression of RFP. This construct, pLac-RBS-RFP, was chosen as our control construct, representing 100% suppression of the engineered frameshift.
 "
"