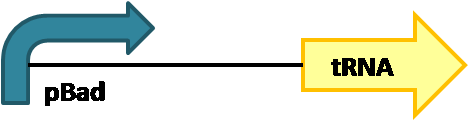Team:MoWestern Davidson/project wetlab
From 2009.igem.org
| Line 1: | Line 1: | ||
{{Template:MoWestern_Davidson2009}} | {{Template:MoWestern_Davidson2009}} | ||
| + | |||
| + | == Completed tRNAs == | ||
We decided to use a weak-medium promoter, pBad, to express our suppressor tRNAs. | We decided to use a weak-medium promoter, pBad, to express our suppressor tRNAs. | ||
| Line 6: | Line 8: | ||
[[Image:Pbad trna.JPG|none|300px]] | [[Image:Pbad trna.JPG|none|300px]] | ||
| - | |||
| - | |||
| - | |||
| - | |||
Revision as of 21:51, 27 July 2009

Completed tRNAs
We decided to use a weak-medium promoter, pBad, to express our suppressor tRNAs.
This promoter choice allows us to induce greater expression with L-arabinose. We found that suppressor tRNAs controlled by pBad is not lethal to E. coli and allows for normal culture growth.
- AGGAC
- AGGAU
- CCAAU
- CCACC
- CCACU
- CCAUC (9 bp-anticodon)
- CCAUC (10 bp-anticodon)
- CCCUC
- CGGUC
- CUACC
- CUACU
- CUAGC
- CUAGU
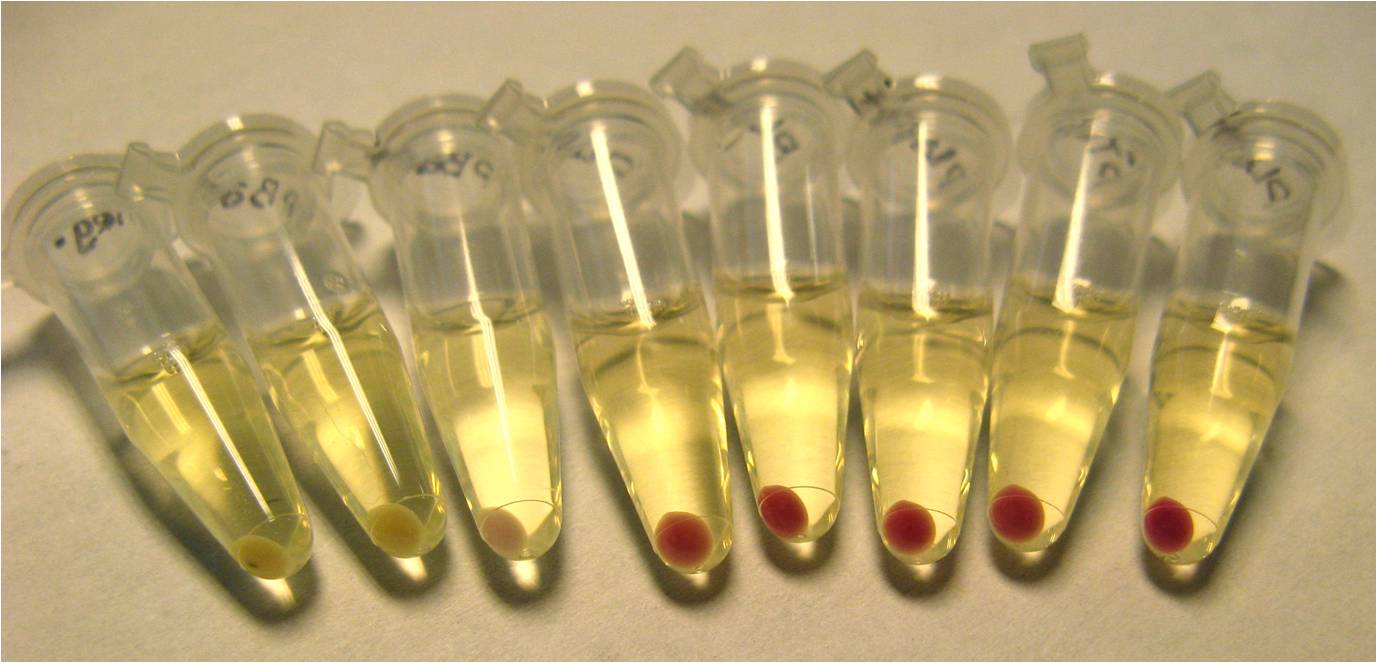
Completed Reporter With Frameshift Suppressor Leaders (FSL)
- CUAGC-Chloramphenicol Acetyltransferase
- CCAUC(9)-Red Fluorescent Protein
- CCAUC(10)-Red Fluorescent Protein
- CCCUC- Red Fluorescent Protein
- CGGUC- Red Fluorescent Protein, tetracycline resistance protein
- CUACU- Red Fluorescent Protein, tetracycline resistance protein
- CUAGU- Red Fluorescent Protein, tetracycline resistance protein
Choosing A Promoter
One important consideration in our project design was choosing the promoter(s) that would induce a notable phenotype. For controlling our suppressor tRNA expression, we wanted a promoter that would transcribe enough tRNA to cause a high probability of suppression but not so much tRNA that cells would become sick. For controlling our reporters with a leading logical clause, we wanted a promoter that would have strong enough transcription to allow more chances for suppression, and thereby more reporter expression.
Promoter Control of Fluorescence Reporters
In order to have a visible expression of RFP or GFP, we constructed parts with RBS-RFP under the control of 4 different commonly-used promoters:
- pBad (Potential induction with L-arabinose)
- pLacI (also called pLac. Potential induction with IPTG)
- pLacIQ (Potential induction with IPTG)
- pTet
We found that pLac induced with IPTG caused the greatest expression of RFP. This construct, pLac-RBS-RFP, was chosen as our control construct, representing 100% suppression of the engineered frameshift.
Promoter Control of Drug Resistance Reporters
 "
"