Team:DTU Denmark/theory
From 2009.igem.org
(New page: image:wiki banner_967px.png <html> <head> <title>Theoretical background</title> <style> #globalWrapper { background-image: "" !important; background-repeat: repeat; } .ideasList{ ...) |
|||
| Line 171: | Line 171: | ||
<font color="" face="arial" size="2"> | <font color="" face="arial" size="2"> | ||
<a href="theory" CLASS=leftbar>- Theoretical background</a><br> | <a href="theory" CLASS=leftbar>- Theoretical background</a><br> | ||
| - | <a href="yeast | + | <a href="yeast" CLASS=leftbar>- Yeast as a model organism</a><br> |
| - | <a href="practicalapproach | + | <a href="practicalapproach" CLASS=leftbar>- Practical approach</a><br> |
<br> | <br> | ||
</font> | </font> | ||
Revision as of 19:17, 8 October 2009

| Home | The Team | The Project | Parts submitted | Modelling | Notebook |
|
The redoxilator - Theoretical background - Yeast as a model organism - Practical approach The USER assembly standard - Principle - Proof of concept - Manual - Primer design software |
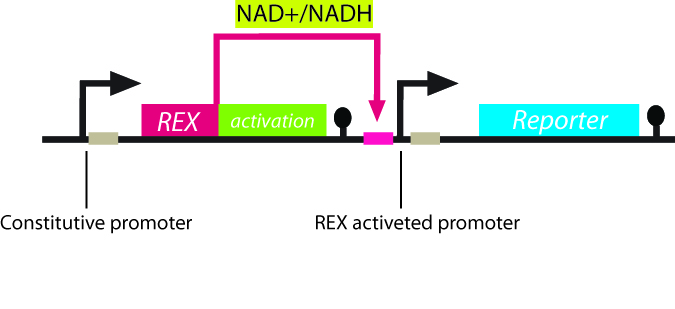
The project Project abstract The Redoxilator By in silico design and computer modelling followed by gene synthesis, we have constructed a molecular NAD/NADH ratio sensing system in Saccharomyces cerevisiae. The sensor works as an inducible transcription factor being active only at certain levels of the NAD/NADH ratios. By the coupling of a yeast optimized fast degradable GFP, the system can be used for in vivo monitoring of NAD/NADH redox poise. A future novel application of the system is heterologous redox coupled protein production in yeast.
The USER fusion standard Another part of our project is the proposal of a new parts-assembly standard for Biobricks based on USER(TradeMark) cloning. With this technique, not based on restriction enzymes, all parts independent of function can be assembled without leaving any scars from the restriction enzyme digestions. |
The yeast metabolic cycle It has recently been shown by Tu et al. and Klevecz et al. that the expression of at least half of the genes monitored on a standard yeast gene chip will oscillate in a coordinated manner when grown under glucose limited conditions. The cells will shift between oxidative and reductive metabolism in a synchronized metabolic cycle with three phases: oxidative, reductive/building and reductive/ charging. As oxygen will only be consumed in the oxidative phase, the dissolved oxygen will oscillate. Many metabolites and cofactors including NADH and NAD+ will also oscillate during this cycle as NADH is converted to NAD+ when oxygen is consumed. |
| Comments or questions to the team? Please Email us -- Comments of questions to webmaster? Please Email us |
 "
"