Team:DTU Denmark/theory
From 2009.igem.org
| Line 195: | Line 195: | ||
<!-- INSERT MAIN TEXT HERE! (formatting: <b>bold</> <i>italic> <h4>header</h4>) --> | <!-- INSERT MAIN TEXT HERE! (formatting: <b>bold</> <i>italic> <h4>header</h4>) --> | ||
| - | <font size="4"><b> | + | <font size="4"><b>Theoretical background</b></font><br><br> |
| - | |||
| - | <p align="justify"> | + | <p align="justify">The NAD<sup>+</sup>/NADH ratio sensor-protein Rex (<u>Re</u>do<u>x</u> regulator) has been discovered in the bacterium Streptomyces |
| + | coelicolor. In its host organism, the sensor works as a repressor and controls the gene expression of a large number of genes by recognizing and binding to a specific DNA-sequence termed ROP (<u>R</u>ex <u>OP</u>erator). NAD<sup>+</sup> and NADH compete for Rex binding, and the protein binds the ROP DNA-sequence only when NAD<sup>+</sup> is bound.</p> | ||
| + | |||
| + | <h4>Our synthetic biology project: The Redoxilator</h4> | ||
| + | <p align="justify">To achieve a system that senses changing levels in the NAD<sup>+</sup>/NADH ratio in the eukaryote <i>S. cerevisiae</i>, the gene encoding the Rex protein will be fused to a yeast activator domain, resulting in a new synthetic protein: the Redoxilator. The ROP sequence - the DNA binding site Rex can bind to - will be inserted into a yeast promoter, resulting in a promoter activated by the Redoxilator.</p> | ||
</html> | </html> | ||
| - | [[Image: | + | [[Image:genedesignA.jpg.redox.jpg|300px|thumb|center|The redox coupled system]] |
<html> | <html> | ||
| + | <br> | ||
| + | <br> | ||
| + | </html> | ||
| + | [[Image:genedesignB.jpg.redox.jpg|300px|thumb|center|The redox coupled system]] | ||
| + | <html> | ||
| + | <br> | ||
| + | |||
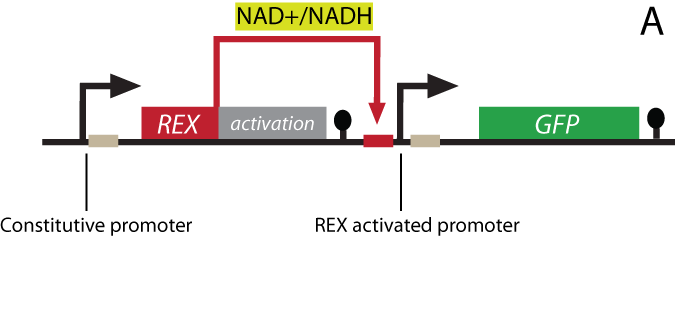
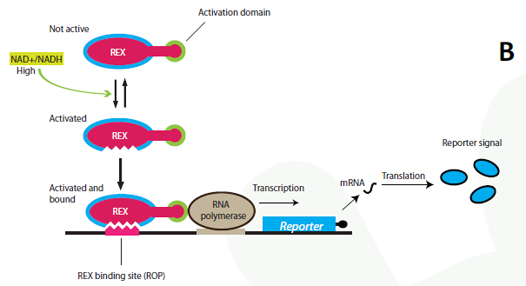
| + | <p align="justify"><i><b>Figure 1 - Gene design and redox regulation</b><br> | ||
| + | <b>A:</b> The Rex gene will be fused to an activator domain forming the Rexivator and will be transcribed constitutively leading to constant concentration of the sensor in the cell. The ROB sequence will be inserted into a specific promoter followed by a reporter | ||
| + | gene, which will only be transcribed if the Rexivator complex is bound to the promoter. <b>B:</b> The Rexivator only binds the ROB DNA sequence under the condition of having NAD+ bound. Under these circumstances the fused activator domain summons the RNA polymerase and the reporter gene will be transcribed</i><p> | ||
| - | < | + | <p align="justify">A certain NAD<sup>+</sup>/NADH ratio will activate the Redoxilator to recognize the ROB promoter resulting in transcription |
| + | of a downstream gene. In this way the ROB promoter and the Redoxilator comprises the complete sensing system. | ||
| + | The system can be coupled to the expression of virtually any gene of interest; making transcription solely dependent | ||
| + | on the ratio of NAD<sup>+</sup>/NADH in the cell. In our iGEM project, the system will be used for two selected | ||
| + | applications considered highly relevant: i) in vivo monitoring of NAD<sup>+</sup>/NADH in yeast, and ii) NAD<sup>+</sup>/NADH ratio | ||
| + | regulated production of yeast products in chemostat processes.</p> | ||
| - | |||
Revision as of 19:21, 8 October 2009

| Home | The Team | The Project | Parts submitted | Modelling | Notebook |
|
The redoxilator - Theoretical background - Yeast as a model organism - Practical approach The USER assembly standard - Principle - Proof of concept - Manual - Primer design software |
The project Theoretical background The NAD+/NADH ratio sensor-protein Rex (Redox regulator) has been discovered in the bacterium Streptomyces coelicolor. In its host organism, the sensor works as a repressor and controls the gene expression of a large number of genes by recognizing and binding to a specific DNA-sequence termed ROP (Rex OPerator). NAD+ and NADH compete for Rex binding, and the protein binds the ROP DNA-sequence only when NAD+ is bound. Our synthetic biology project: The RedoxilatorTo achieve a system that senses changing levels in the NAD+/NADH ratio in the eukaryote S. cerevisiae, the gene encoding the Rex protein will be fused to a yeast activator domain, resulting in a new synthetic protein: the Redoxilator. The ROP sequence - the DNA binding site Rex can bind to - will be inserted into a yeast promoter, resulting in a promoter activated by the Redoxilator.
Figure 1 - Gene design and redox regulation
A certain NAD+/NADH ratio will activate the Redoxilator to recognize the ROB promoter resulting in transcription of a downstream gene. In this way the ROB promoter and the Redoxilator comprises the complete sensing system. The system can be coupled to the expression of virtually any gene of interest; making transcription solely dependent on the ratio of NAD+/NADH in the cell. In our iGEM project, the system will be used for two selected applications considered highly relevant: i) in vivo monitoring of NAD+/NADH in yeast, and ii) NAD+/NADH ratio regulated production of yeast products in chemostat processes. |
The yeast metabolic cycle It has recently been shown by Tu et al. and Klevecz et al. that the expression of at least half of the genes monitored on a standard yeast gene chip will oscillate in a coordinated manner when grown under glucose limited conditions. The cells will shift between oxidative and reductive metabolism in a synchronized metabolic cycle with three phases: oxidative, reductive/building and reductive/ charging. As oxygen will only be consumed in the oxidative phase, the dissolved oxygen will oscillate. Many metabolites and cofactors including NADH and NAD+ will also oscillate during this cycle as NADH is converted to NAD+ when oxygen is consumed. |
| Comments or questions to the team? Please Email us -- Comments of questions to webmaster? Please Email us |
 "
"