Team:DTU Denmark/modelling
From 2009.igem.org

| Home | The Team | The Project | Parts submitted | Modelling | Notebook |
|
Also check out our Danish website - DTU iGEM 2009 |
Modelling of GFP expression Introduction Here a model for the GFP expression under the control of “The Redoxilator” expression system is derived. The model links the level of the signal molecules NADH and NAD to the final expression of GFP through the events of transcription factor activation, promoter activation, synthesis and degradation of mRNA and finally synthesis, dilution and degradation of GFP.NADH and NAD binding to the transcription factor Both NADH and NAD binds reversible to the same site of the transcription factor (TF):Dissociation of TF and NADH at equilibrium: Dissociation of TF and NAD: Dissociation of TF and NAD: Balance of N and H. Constant total level of NADH + NAD in the cell are assumed: Balance of TF. TF is constitutively expressed leading to constant total value N inhibits DNA binding activity of TF and thus transcription activity. Active TF is designated TF’. Leading to the conserved moiety: Using (3), the fraction of TF’ (tf) can be written as With (1) and (2) in (4) the tf becomes  Leading to Graph of with different changing values of H/N. Figure 1: Transcription factor activity with increasing NADH/NAD ratio is hyperbolic. At NADH/NAD = 0.1, tf = 0.5, i.e. half of the transcription factors are active. (For this plot the following parameters were used: K_H = 2 μM, assumed, no experimental value available. K_N = 0,02 μM, NADH affinity > NAD affinity [2]. Promoter binding of active transcription factor When the active TF (TF') is bound to the promoter P, P becomes active (P’) and transcription is initiated. The number of TF needed for promoter activation is designated m:Here it assumed that binding of TF and TFN is equal. The dissociation equilibrium for this reaction: 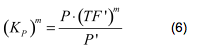 The number of promoters is constant, and hence the following sum of fractions is true Where the fraction of active promoter Pf is Introducing P’ from (6) gives 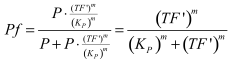 Now introducing  (For simplicity the expression of (5) has not been inserted). Figure 2: Fraction of active promoters with increasing NADH/NAD ratio. The curve is sigmoidal and has almost switch like behavior. From NADH/NAD = 0 to 0.01 the difference in active promoter is almost 60 %.(Assumed value of K_P = 1 mM). mRNA production mRNA is produced from the transcription of the active promoter. Concentration of intracellular levels of mRNA (Here it is assumed that mRNA is degraded corresponding to a first order reaction and that mRNA is not exported from the cell. The term Production rate of mRNA It is assumed that when RNA polymerase is efficiently bound to the promoter, the promoter is active and mRNA is transcribed at constant maximum rate For a given organism Steady state concentration of mRNA At steady state (11) is equal to zero since no mRNA accumulates. Here the mRNA concentration is: Figure 3: mRNA levels at steady state with increasing NADH/NAD ratios. Dilution is neglected since degradation rate of mRNA is much larger. (Assumed parameters: kdm = 8.3 h-1 (half life = 5 min), Np = 60 (2 micron plasmid is used). and GFP production Finally, the mRNA is used as template for the process of translation to protein, here GFP. The GFP protein balance:As with mRNA it is assumed that GFP is degraded by first order rate. The dilution cannot be neglected since protein degradation is much slower than mRNA degradation. Furthermore it is assumed that GFP is not transported out of the cell. The protein is synthesized by ribosomes. The number of ribosomes bound to one mRNA (Np) and the protein production rate of the ribosome (kp) should be taken into account. The number of ribosomes in the cell, ATP and available charged amino acids are assumed to be non-limiting. Taking this into account the protein synthesis rate can be described as Making the final GFP balance Looking back, we now have the GFP balance only depending on the levels of NADH and NAD (through mRNA level, promoter activity and transcription factor activity). Steady state concentration of GFP From (15) we find that the steady state concentration of GFP is  Figure 4: Steady state GFP levels as a direct function of NADH/NAD signal levels. This model tells the metabolic state (through NADH/NAD ratio) of a cell in steady state. (Assumed parameters Nrib = 1. kp = 288 U/h (assuming 40 amino acids per second) k_dGFP = 1.386 h-1 (half time = 30 min)). The dynamic state When the cells are exposed to a certain NADH/NAD level mRNA and in turn GFP levels will change in time according to their balances. In figure XX the NADH/NAD ratio is changed from XX (resting state) to a more stressed state where NADH/NAD = XX. If the NADH/NAD level changes in time all values depending on NADH and NAD will also be time depending. In the yeast metabolic cycle NADH/NAD ratio oscillates in time, resulting in oscillation-production of the protein under control of the redox system:References [1] Heijnen J.J 2009. Modular structured kinetic models: protein production from genes. [2] Mark and Paget, 2003. A novel sensor of NADH/NAD…. [3] Mateus and Avery, 2000. Destabilized green fluorescent protein for monitoring dynamic changes in yeast gene expression with flow cytometry. Yeast 2000; 16: 1313±1323. |
Modelling facts Abbreviations used in the model TF = the transcription factor based on the REX protein TF' = active transcription factor N = concentration of NAD H = concentration of NADH TFH = concentration of the transcription factor bound to NADH. TFN = concentration of the transcription factor bound to NAD. P = Promoter upstream of GFP P' = activated/transcribed promoter tf = fraction of active transcription factor Pf = fraction of active promoter |
| Comments or questions to the team? Please Email us -- Comments of questions to webmaster? Please Email us |
 "
"




