Team:LCG-UNAM-Mexico/LauraJournal
From 2009.igem.org
Goal and objectives
At the start I was working with the delivery system which includes P4 and P2 bacteriophages. However, on August I focused my attention on the phages involved in the defense system, T7 and T3.
My main goals were:
- To obtain all the relevant experimental information about T7 and T3 as the burst sizes and the growth plots with and without phage.
- To generate data to feedback the infection model.
I used two main protocols the Purification of T7 from 35ml lysates from the T7Select System Manual and the Titering Protocol described in the same Manual.
The activities ordered chronologically are described next.
August
On August I was working in verify the accurate of these protocols and in select a strain for the following experiments. In this month a purification protocol was realized with Escherichia coli K-12 strain, the lysis was unsuccessful. To probe the activity of the phague stocks I get the BL21 strain to work with T3 and T7, a purification protocol was realized with Escherichia coli BL21 strain, the lysis was successful. T3 and T7 bacteriophagues stocks were active. It was decided to use C1a to work with T3 and T7 because it is a K-12 derivative and because the next experiments will be focused on this strain. The purification of T7 and T3 using C1a strain was successful (OR stock)
In this month all the cultures get contaminated with yeast (strains C331, C-117, C1a, C-1906, c520, c1895, c2423). The contaminated cultures were transfered to generate spectomycin resistant. Resistants strains c-117, c1a, c331 and c1906 were obtained. I did PCR’s to be sure of the genome integrity. I amplified Cox for c117, c1a and c331; Ogr for c117, c331 and c1a and P4 for c1906, c1a and c331. I charged an agarose gel to check the PCRs, cox and ogr were seen when they were supposed to be. The resistant strains were contaminated with yeast. So, I did no more efforts in this. New stock of the main C1a stock was obtained.
September
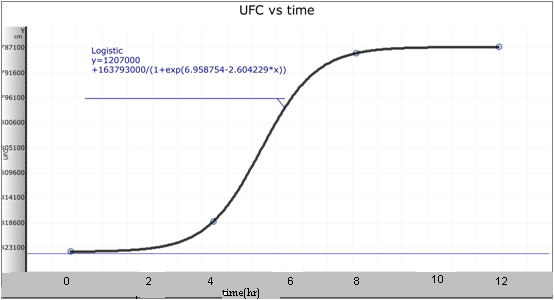
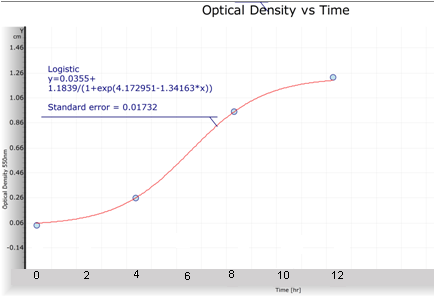
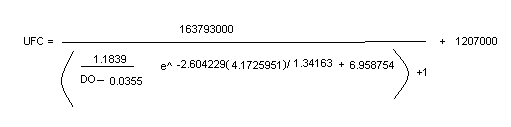
From a culture with OD=0.8 I transferred 1.25ml to have a 100ml culture with OD=0.01. Every four hours for a period of 12 hours I took 1ml from this culture to take an OD measure(to 550nm) and I took another ml to do dilutions, I did cultures corresponding to the 10^-4, 10^-5 and 10^-6 dilutions, and an additional 10^-3 dilution for the first measure. I counted the number of UFCs (unit forming colonies) for these cultures and I calculated the number of UFC per ml in the original culture at time in which the sample was taken. After this, I obtained the formulae with the best adjustment to the points. The corresponding plots and formulae are presented next.
With both plots I obtained a relation between OD and UFC.
The purification protocol was repeated three times for each phague and every stock (1BS, 2BS and 3BS) was tittered including the OR stock. I always follow the of T7 from 35ml lysates from the T7Select System Manual protocol and the tittering protocol of the same manual. Here I present some important details about these experiments.
For the first purification (1BS stock) I mixed 5ul of T7 OR stock with 34ml ofM9LB plus c1a(DO 0.84) and 5ul of T3 OR stock with 33ml of M9LB plus c1a (OD 0.85). It was left by three hours to reach a total lysis. For the tittering of this stock I did dilutions, I took 100ul of the dilution and I mixed it with 350ml of cells and with 3ml of top agar. In the next purifications I only change the volume and the OD of the culture. For the second purification it was 32ml with 0.77OD for both, T7 and T3. For the third purification it was 34ml for both bacteriophagues with a OD=0.94 for T3 and OD=0.94 for T7. The tittering for each stock was repeated some times, the results are presented in the next tables. The non congruent experiments between the dilutions are not presented here.
The number of plaques obtained for each experiment are shown in the next tables ( the numbers after the name of the stock indicate the number if the repetition)
|
DILUTION |
OR1 |
OR2 |
OR3 |
|
10-8 |
167 |
76 |
50 |
|
10-9 |
16 |
7 |
5 |
|
10-10 |
2 |
0 |
0 |
|
DILUTION |
1BS1 |
1BS2 |
2BS1 |
2BS2 |
3BS1 |
3BS2 |
3BS3 |
|
10-9 |
417 |
210 |
NC |
NC |
Clear |
Clear |
Clear |
|
10-10 |
37 |
25 |
NC |
NC |
Clear |
Clear |
Clear |
|
10-11 |
4 |
3 |
480 |
480 |
NC |
Clear |
Clear |
|
10-12 |
--- |
--- |
---- |
--- |
364 |
NC |
NC |
NC=non-countable, Clear = all the cells were dead, ---=the experiment wasn’t done.
T3
|
DILUTION |
OR1 |
OR2 |
1BS1 |
1BS2 |
1BS3 |
IBS4 |
2BS1 |
2BS2 |
2BS3 |
|
10-8 |
350 |
104 |
NC |
Clear |
Clear |
Clear |
NC |
NC |
Clear |
|
10-9 |
30 |
10 |
210 |
Clear |
NC |
Clear |
480 |
182 |
NC |
|
10-10 |
3 |
0 |
25 |
NC |
254 |
NC |
63 |
23 |
600 |
|
10-11 |
--- |
--- |
3 |
250 |
25 |
251 |
8 |
2 |
74 |
The same nomenclature
The results were conclusive for
T7 OR stock: 1x108 UFP/ul
T7 1BS: 3.72x1012 UFP totales
T7 2BS:5.76x1014 UFP totales
T73BS: one or two magnitude order above the last results.
T3 OR: 2x108
BURST SIZE OBTAINED WITH PURIFICATION DATA.
These results were used to obtain the “burst size” of T7 bacteriophagues. I supposed some things: all the phagues are infecting a bacterium, all the cells were infected and almost all the bacteria died in the first round of infection. I calculated the bacteria number with the formula obtained with the growth curve which involves OD and UFC. So I calculate the burst size as the total number of phagues divided by the total number of bacteria. The data of the first purification (1BS stock )indicate a burst size of 952 phagues per bacterium. However the data of the 2BS stock indicate a burst size of 160,000 phagues per bacterium and surprisingly the data of the 3BS stock indicate a bigger number. We now it is not possible because the phague production is limited by the amount of nucleotides available. For this reason, I designed other experiment to measure the burst size.
October
I did a growth curve with phague.
In the first assay I cultured 35ml of M9LB c1a when the OD reached a value close to 0.8 I add 5ul of T3 or T7. I took OD measures every twenty minutes. No phague was added in the control. The data recollected for this experiment is shown in the next table. I did the assay twice. Because I took different initial values for this replicate and because I didn’t plates I repeated the assay taking into account these facts.
 "
"