Team:Todai-Tokyo/Notebook/isoleucine
From 2009.igem.org
Contents |
Plan
Aim: Create bacteria that produce a noxious odour when induced to do so
Methods:
- Clone the yqiT gene from Bacillus subilitis into a biobrick vector.
- Make constructs that express this gene constitutively and under the control of an inducible promoter.
6/5
Constructs to be created:
- yqiT
- ptetR-RBS-yqiT-dterm
- pAraC-RBS-yqiT-dterm
- pLacI-RBS-yqiT-dterm
Obtaining DNA:
Resuspended DNA in the following wells with 10ul water:
Plate 1 1D
[http://partsregistry.org/wiki/index.php/Part:BBa_R0080 AraC regulated promoter]
Plate 1 12E
[http://partsregistry.org/wiki/index.php?title=Part:BBa_R0010 LacI regulated promoter]
Plate 1 1H
[http://partsregistry.org/wiki/index.php/Part:BBa_B0030 Strong RBS]
Plate 1 13B
[http://partsregistry.org/wiki/index.php?title=Part:BBa_J13002 TetR repressed POPS generator]
Plate 2 24C
[http://partsregistry.org/wiki/index.php?title=Part:BBa_B0014 Double terminator]
Transformed 1ul of each of the above into DH5a competent cells:
Transformation
- Mix 1ul of DNA with 100ul of competent cells on ice.
- Leave on ice for 30 minutes.
- Heat shock at 42℃ for 45 seconds.
- Leave on ice for 2 minutes.
- Add 500ul of LB and incubate at 37℃ for 1 hour.
- Plate on LB-ampicillin plates.
6/6
No colonies grew from the 5 transformations of 6/5.
6/7
Creating a biobrick part out of yqiT
Strategy: PCR out the yqiT gene from the Bacillus subilitis genome using primers to attach the biobrick preffix/suffix and clone this into a biobrick vector. The vector utilized is the GFP generator which houses a sufficient-sized insert:
Plate 1 16E (from 2007 iGEM distribution provided by Chiba University)
[http://partsregistry.org/wiki/index.php?title=Part:BBa_E0840 GFP generator]
PCR of yqiT gene
The Bacillus subilitis genome was provided by <?????????>
The following primers were used to amplify an approx. 1500bp fragment from the Bacillus subilitis genome.
yqiT EX: <????????>
yqiT SP: <????????>
PCR protocol
The following was mixed in a PCR tube:
1ul 20uM yqiT EX primer
1ul 20uM yqiT SP primer
2ul 10X <????> buffer
<???> dNTP mix
0.15ul Bacillus subilitis genome
<???> <????> enzyme
<???> MilliQ
Performed PCR using the following program:
1. 95℃ 2 minutes
2. 95℃ 1 minute
3. 50℃ 30 seconds
4. 72℃ 1 minute 20 seconds
5. Repeat 2-4 29 times
6. 4℃ ∞
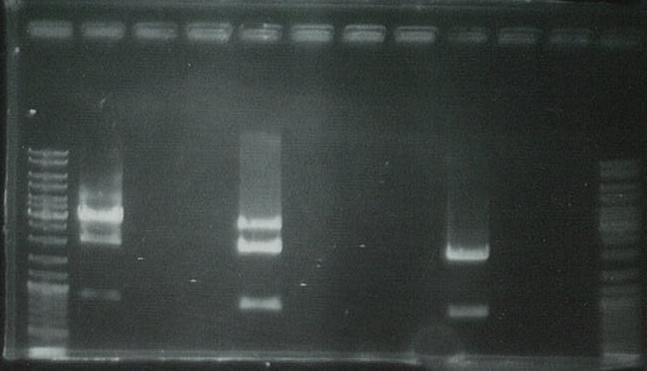
Purified PCR product using the Promega PCR purification kit.
Ran on gel to visualize bands:
Digestion
Digested purified PCR product and the GFP generator both with XbaI/PstI:
yqiT
3ul PCR product
1ul High buffer
0.5ul XbaI
0.5ul PstI
5ul MilliQ
GFP generator
6ul DNA
2ul High buffer
0.5ul XbaI
0.5ul PstI
11ul MilliQ
Incubated mixtures at 37℃ for 1 hour.
Ran on gel to gel purify:
 "
"