Team:Todai-Tokyo/Notebook/isoleucine
From 2009.igem.org
Contents |
Plan
Aim: Create bacteria that produce a noxious odour when induced to do so
Methods:
- Clone the yqiT gene from Bacillus subilitis into a biobrick vector.
- Make constructs that express this gene constitutively and under the control of an inducible promoter.
6/5
Constructs to be created:
- yqiT
- ptetR-RBS-yqiT-dterm
- pAraC-RBS-yqiT-dterm
- pLacI-RBS-yqiT-dterm
Obtaining DNA:
Resuspended DNA in the following wells with 10ul water:
Plate 1 1D
[http://partsregistry.org/wiki/index.php/Part:BBa_R0080 AraC regulated promoter]
Plate 1 12E
[http://partsregistry.org/wiki/index.php?title=Part:BBa_R0010 LacI regulated promoter]
Plate 1 1H
[http://partsregistry.org/wiki/index.php/Part:BBa_B0030 Strong RBS]
Plate 1 13B
[http://partsregistry.org/wiki/index.php?title=Part:BBa_J13002 TetR repressed POPS generator]
Plate 2 24C
[http://partsregistry.org/wiki/index.php?title=Part:BBa_B0014 Double terminator]
Transformed 1ul of each of the above into DH5a competent cells:
Transformation
- Mix 1ul of DNA with 100ul of competent cells on ice.
- Leave on ice for 30 minutes.
- Heat shock at 42℃ for 45 seconds.
- Leave on ice for 2 minutes.
- Add 500ul of LB and incubate at 37℃ for 1 hour.
- Plate on LB-ampicillin plates.
6/6
No colonies grew from the 5 transformations of 6/5.
6/7
Creating a biobrick part out of yqiT
Strategy: PCR out the yqiT gene from the Bacillus subilitis genome using primers to attach the biobrick preffix/suffix and clone this into a biobrick vector. The vector utilized is the GFP generator which houses a sufficient-sized insert:
Plate 1 16E (from 2007 iGEM distribution provided by Chiba University)
[http://partsregistry.org/wiki/index.php?title=Part:BBa_E0840 GFP generator]
Overview:
- PCR yqiT gene from genome
- purify DNA from the PCR product
- digest GFP generator and purified PCR product by XbaI/PstI
- gel-purify the digested vector (GFP generator without the insert i.e. empty) and the yqiT gene
- ligate the above 2 fragments of DNA together
- transform into E. coli and select for ampicillin resistance
- check for transformation success using colony PCR by yqiT primers
PCR of yqiT gene
The Bacillus subilitis genome was provided by <?????????>
The following primers were used to amplify an approx. 1500bp fragment from the Bacillus subilitis genome.
yqiT EX: <????????>
yqiT SP: <????????>
PCR protocol
The following was mixed in a PCR tube:
1ul 20uM yqiT EX primer
1ul 20uM yqiT SP primer
2ul 10X <????> buffer
<???> dNTP mix
0.15ul Bacillus subilitis genome
<???> <????> enzyme
<???> MilliQ
Performed PCR using the following program:
1. 95℃ 2 minutes
2. 95℃ 1 minute
3. 50℃ 30 seconds
4. 72℃ 1 minute 20 seconds
5. Repeat 2-4 29 times
6. 4℃ ∞
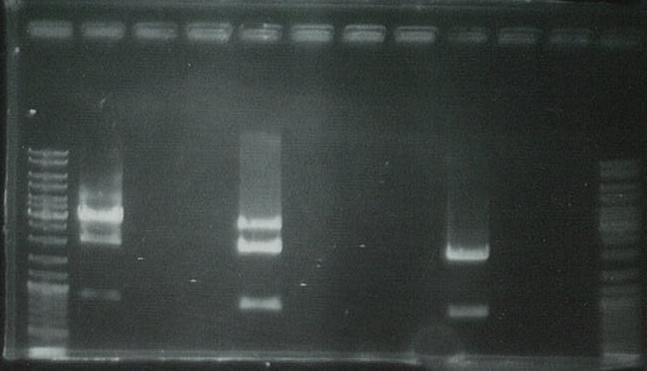
Purified PCR product using the Promega PCR purification kit.
Ran on gel to visualize bands:
PCR unsuccessful: will digest the GFP generator but cannot perform the ligation without the PCR product.
Digestion
Digested purified PCR product and the GFP generator both with XbaI/PstI:
yqiT
3ul PCR product
1ul High buffer
0.5ul XbaI
0.5ul PstI
5ul MilliQ
GFP generator
6ul DNA
2ul High buffer
0.5ul XbaI
0.5ul PstI
11ul MilliQ
Incubated mixtures at 37℃ for 1 hour.
Ran on gel to gel purify:
Lanes 1, 3, 5:
Lanes 2, 4, 6:
Cut out the second band from the bottom and dissolved the gel using the Promega gel purification kit to extract DNA from.
This DNA is stored for later usage as the GFP generator X/P vector fragment.
 "
"