EPF-Lausanne/21 August 2009
From 2009.igem.org
Contents |
Wet Lab
Miniprep of the 2 possible readout 1 plasmids.
Colony PCR of the LacI-RBS-LovTap-Term that grew to check whether the insert is correct. Then agarose gel to verifiy length of fragment.
Also, since we wanted to do further tests on the maybe readout 1 plasmids, we did a classic PCR with the iGEM primers on the plasmids, and a digestion assay with SpeI (since the TrpOperon has 2 SpeI sites in its sequence). Checked the results on the gel as well.
Results of the first characterization
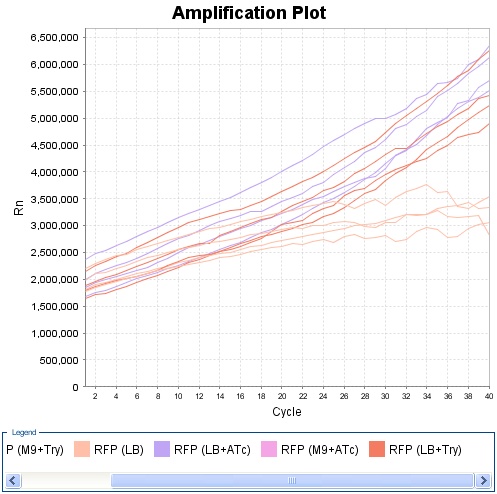
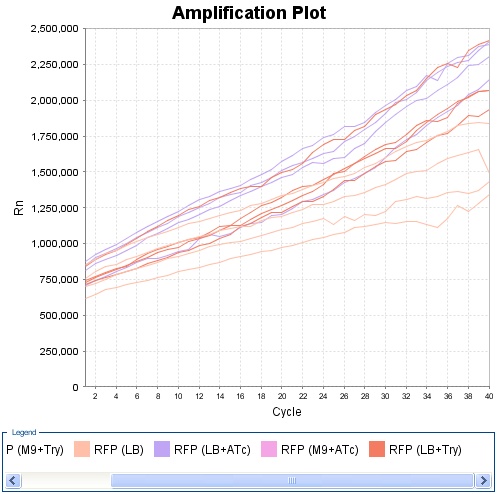
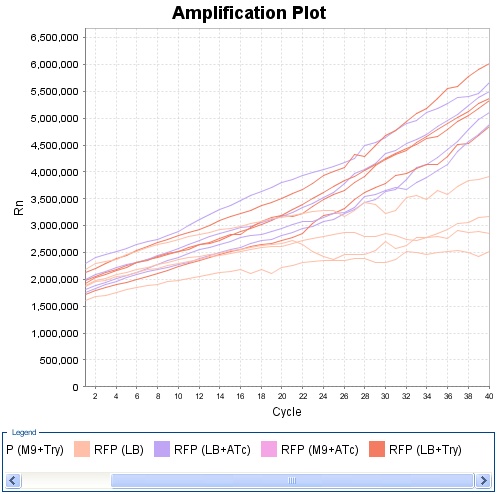
Experiment on Read Out 2. Only the clones 4, 5 and 10 had nice curves. The experiment was made using LB. For every clone there was three different possibilities : without anything OR with tryptophan OR with ATC. The cells had all exactly the same initial conditions, the products were added only at the last moment. We can see that clone 4 and 10 show a very good answer to ATC as well as to TRP.
Clone 4 :
Clone 5 :
Clone 10 :
Processing of the data
As we had 4 wells at each same condition, we made a mean for each condition for the clones #4 and #10. Then an interessting thing to do is the relative difference between the response to Try or ATc and the signal of the negative control.
Clone #4:
Clone #10:
If we try to fit the plot of the relative difference e.g. the effective response of our system, we see that a polynomial function of second order fit the best. Normally we expect to have an exponential response, so we will need to do more experiments.
People in the lab
Basile, Gab, Christian


 "
"