Construction of pRhaFlp
From 2009.igem.org
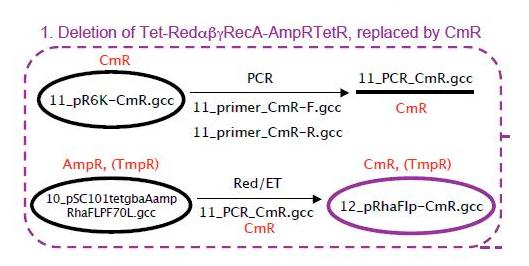
Deletion of Rha-Red alpha beta gamma RecA-AmpRTetR, replaced by CmR
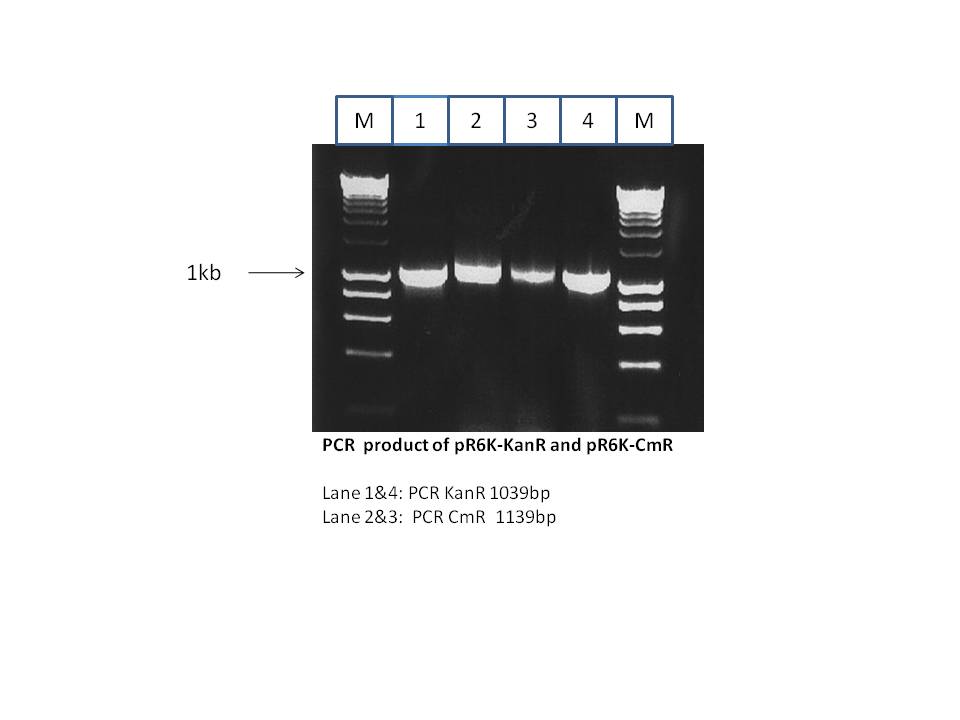
As shown above the CmR gene which encodes for chloroamphinicol resistance was amplified from the pR6K-CmR plasmid (from Stewart lab). This CmR gene has homology arms to enable its introduction into the pRhaFlp plasmid (from Stewart lab) by Red/ET recombination. The pRhaFlp CmR has a pSC101 temperature sensitive origin (grows at 30°C but not at 37°C). The PCR product was isolated and purified from the gel. The gel was as shown below
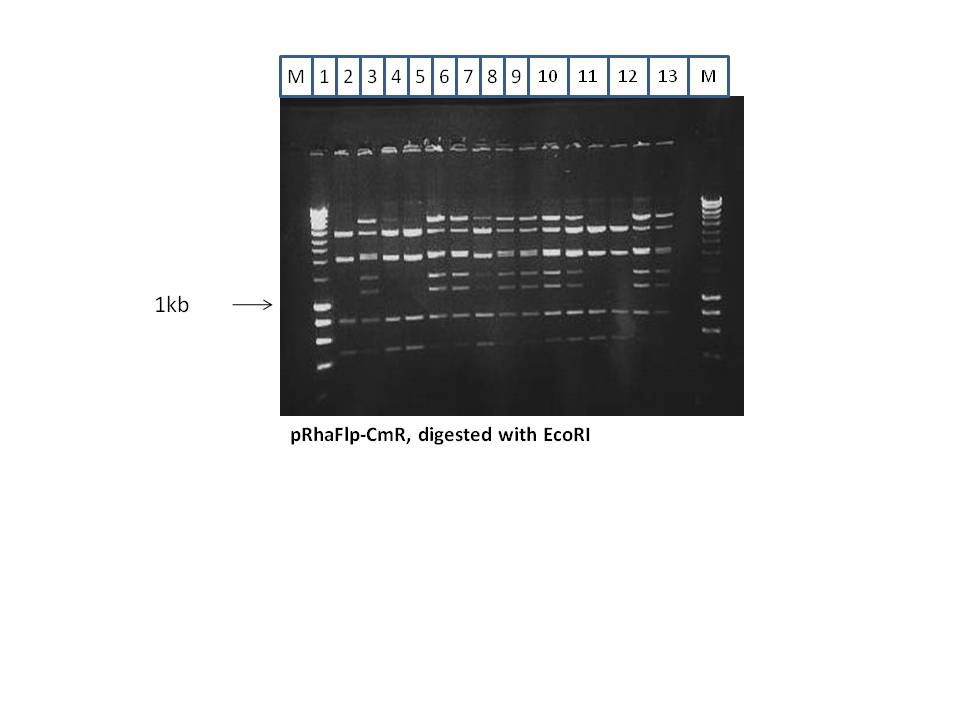
Red/ET was done to introduce the CmR gene into the pRhaFlp. The resulting plasmid was named pRhaFlp CmR. This was digested with EcoRI, and showed the following results
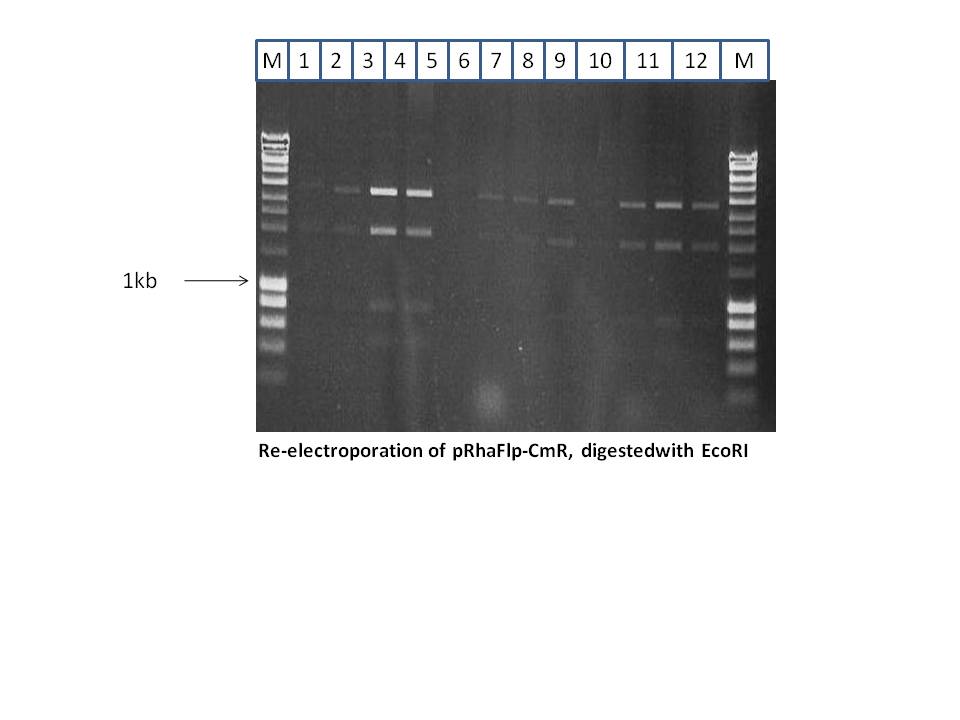
From the 13 minis, 3 correct clones were re-electroporated, in order to purify it. This is a high copy plasmid and the first step of Red/Et mostly results in a mixture of the original plasmid and the desired plasmid. Re-electroporation and transformation results in purer products. 12 minis were done as shown below
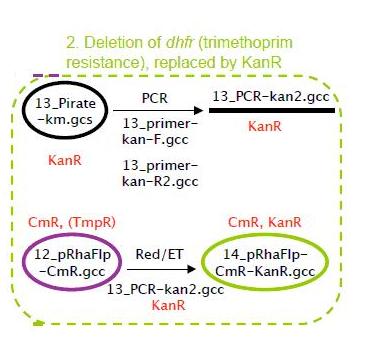
Deletion of dhfr(trimethoprim resistance),replaced by KanR
The dhfr gene encodes for trimetoprim resistance. This resistance is required at a later stage of the project (in the fosmid), and to avoid interference the dhfr gene was deleted from the pRhaFlp plasmid. This was replaced by the KanR gene, as shown in the strategy above. Red/ET was done to delete the dhfr gene and replace it by KanR. The resulting plasmid minis were digested and we got 4 correct clones, as shown below
This was re-electroporated and transformed. 25 minis were digested as shown below and 4 correct clones were kept for further use.
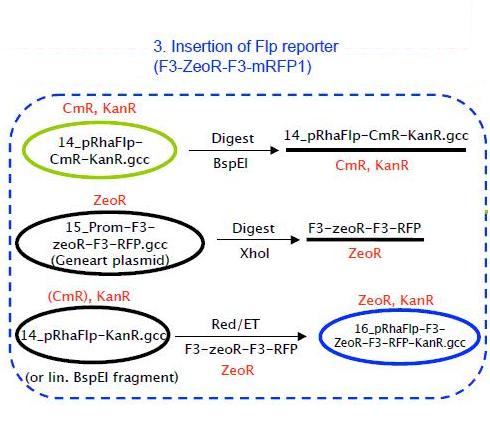
Insertion of Flp reporter
As shown in the strategy above, pRhaFlp CmR was digested using BspEI in order to linearize it.
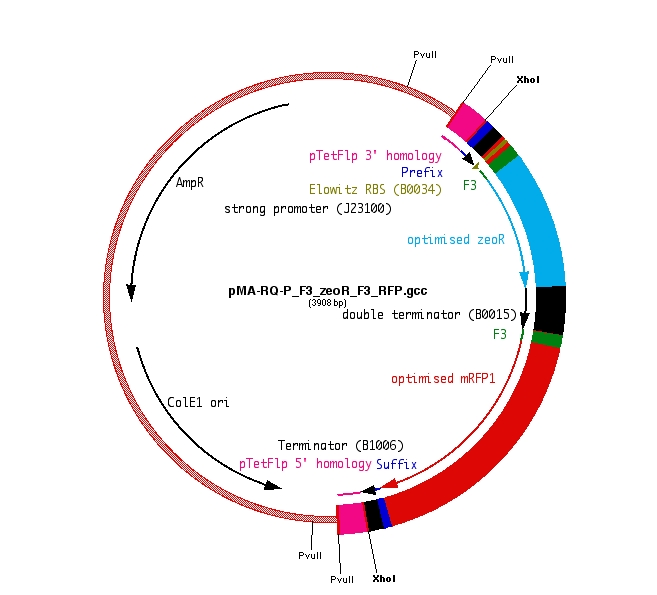
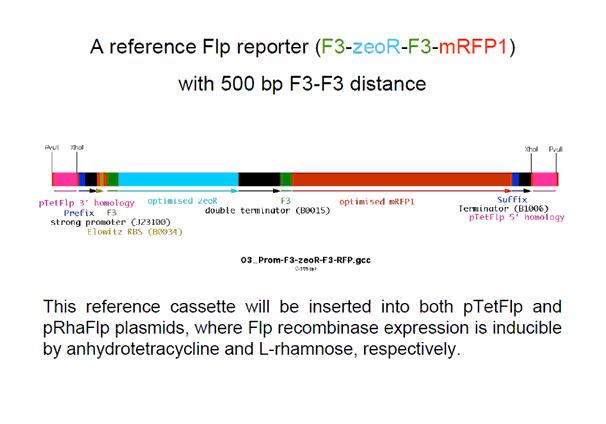
F3-ZeoR-F3-RFP (plasmid ordered from GeneArt), is as shown in Fig 8. This when digested using PvuII resulted in the fragment shown in Fig 9. It carries a zeocin resistance gene (ZeoR). The ZeoR gene is flanked by F3 sites that are similar to the FRT sites (vary in a few bases) and is hence recognized and excised by Flippase (Flp). There is a terminator upstream of the second F3 site and a RFP gene is immediately downstream. So in the absence of Flp RFP is not expressed, But when Flp excises the region between the F3 sites, RFP is expressed.
The PvuII digest was as shown below.
From this the band of size 1549 bp was isolated from the gel and purified.
Gel pic 11
Both pRhaFlp KanR and F3-ZeoR-F3-RFP fragments were linear, so linear+linear Red/ET was done in GB05-DIR cells (from Stewart lab).
 "
"