Team:Bologna/Project
From 2009.igem.org
| (21 intermediate revisions not shown) | |||
| Line 18: | Line 18: | ||
Transcription of the target gene yields a mRNA strand - containing the CIS-repressing sequence at its 5' end - available for translation into protein by ribosomes (<i>see Fig. 1, left panel</i>). When the promoter controlling the TRANS coding sequence is active, it drives the transcription of an oligoribonucleotide complementary to the CIS mRNA sequence. The TRANS/CIS <b>RNA duplex</b> prevents ribosomes from binding to RBS on target mRNA, thus <b>silencing protein synthesis</b>. The amount of the TRANS-repressor regulates the rate of translation of the target mRNA (<i>see Fig. 1, right panel</i>) | Transcription of the target gene yields a mRNA strand - containing the CIS-repressing sequence at its 5' end - available for translation into protein by ribosomes (<i>see Fig. 1, left panel</i>). When the promoter controlling the TRANS coding sequence is active, it drives the transcription of an oligoribonucleotide complementary to the CIS mRNA sequence. The TRANS/CIS <b>RNA duplex</b> prevents ribosomes from binding to RBS on target mRNA, thus <b>silencing protein synthesis</b>. The amount of the TRANS-repressor regulates the rate of translation of the target mRNA (<i>see Fig. 1, right panel</i>) | ||
</html> | </html> | ||
| - | + | <br><br> | |
[[Image:project3b.png|center|950px|thumb|<center>Figure 1 - T-REX device</center>]] | [[Image:project3b.png|center|950px|thumb|<center>Figure 1 - T-REX device</center>]] | ||
<br><br> | <br><br> | ||
| - | <font face="Calibri" font size="5" color="#000000"><b>CIS and | + | <font face="Calibri" font size="5" color="#000000"><b>CIS and TRANS Parts Design</b> |
<br><br> | <br><br> | ||
<font face="Calibri" font size="4" color="#000000"> | <font face="Calibri" font size="4" color="#000000"> | ||
<html> | <html> | ||
<font face="Calibri" font size="4" color="#000000"> | <font face="Calibri" font size="4" color="#000000"> | ||
| - | To identify CIS-repressing and TRANS-repressor | + | To identify CIS-repressing and TRANS-repressor DNA sequences, we developed <a href="https://2009.igem.org/Team:Bologna/Software">BASER</a> software. We used it to seek for two complementary 50bp non-coding sequences, whose transcribed RNAs:<br> |
a) feature maximal free energy in the secondary structure (i.e. reducing the probability of its intra-molecular annealing); <br> | a) feature maximal free energy in the secondary structure (i.e. reducing the probability of its intra-molecular annealing); <br> | ||
b) have minimal unwanted interactions with genomic mRNA; <br> | b) have minimal unwanted interactions with genomic mRNA; <br> | ||
c) present a minimal probability of partial/shifted hybridization with complementary strands. <br><br> | c) present a minimal probability of partial/shifted hybridization with complementary strands. <br><br> | ||
| - | Here below are the CIS-repressing and TRANS-repressor sequences: | + | Here below are the CIS-repressing and TRANS-repressor sequences. More details about BASER and its functioning can be found in the <html><a href="https://2009.igem.org/Team:Bologna/Software">software section</a>.</html> |
<br><br> | <br><br> | ||
| - | |||
{| align="center" | {| align="center" | ||
| Line 79: | Line 78: | ||
|} | |} | ||
<br><br> | <br><br> | ||
| - | |||
| - | |||
| - | |||
<html> | <html> | ||
<font face="Calibri" font size="5" color="#000000"><b>Testing Circuit</b> | <font face="Calibri" font size="5" color="#000000"><b>Testing Circuit</b> | ||
| Line 91: | Line 87: | ||
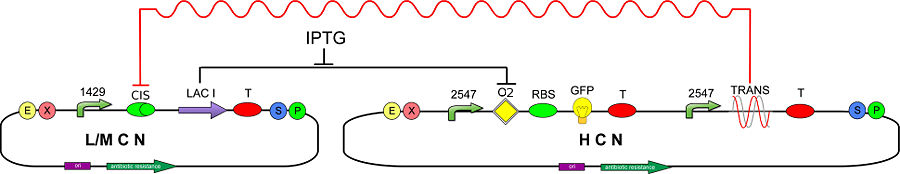
[[Image:circuit2OK.jpg|center|900px|thumb|<center>Figure 2 - Genetic Circuit to test CIS and TRANS' mRNA functionality</center>]] | [[Image:circuit2OK.jpg|center|900px|thumb|<center>Figure 2 - Genetic Circuit to test CIS and TRANS' mRNA functionality</center>]] | ||
<br> | <br> | ||
| - | The CIS-repressing sequence is assembled upstream of | + | The CIS-repressing sequence is assembled upstream of <i>lac</i> I (BBa_C0012), therefore the synthesis of LacI should be silenced/damped by the constitutively transcribed TRANS-repressor mRNA. To detect silencing of LacI, due to the action of T-REX, we realized a new inverter (BBa_K201001) consisting of a promoter regulated by LacI (BBa_K201008) and a GFP reporter (BBa_J04031).<br> |
We expect that a TRANS-repressor oligoribonucleotide with high affinity to CIS-repressing mRNA, inhibits the translation of LacI and then determines a maximally expressed GFP. Otherwise, in case of low TRANS/CIS affinity one should expect partially (or completely) repressed GFP expression.<br> | We expect that a TRANS-repressor oligoribonucleotide with high affinity to CIS-repressing mRNA, inhibits the translation of LacI and then determines a maximally expressed GFP. Otherwise, in case of low TRANS/CIS affinity one should expect partially (or completely) repressed GFP expression.<br> | ||
| - | To maximize the probability to silence the CIS transcript and switch on the GFP, we decided to use a high copy number (HCN) plasmid (pSB1A2) for the TRANS-repressor and a low copy number (LCN) plasmid (pSB3K3) for the LacI generator. <br>If the GFP inverter is unable to reveal the LacI reduction due to T-REX action, because of a high level of the free LacI concentration, IPTG can be | + | To maximize the probability to silence the CIS transcript and switch on the GFP, we decided to use a high copy number (HCN) plasmid (pSB1A2) for the TRANS-repressor and a low copy number (LCN) plasmid (pSB3K3) for the LacI generator. <br>If the GFP inverter is unable to reveal the LacI reduction due to T-REX action, because of a high level of the free LacI concentration, IPTG can be supplied to reduce free LacI. In fact, the sensitivity of the GFP inverter to LacI variations depends on free LacI concentration. Using IPTG is thus possible to set actual LacI value in the region where the inverter has the highest sensitivity. |
<br><br> | <br><br> | ||
<font face="Calibri" font size="5" color="#000000"><b>Mathematical Model</b> | <font face="Calibri" font size="5" color="#000000"><b>Mathematical Model</b> | ||
<br><br> | <br><br> | ||
<font face="Calibri" font size="4" color="#000000"> | <font face="Calibri" font size="4" color="#000000"> | ||
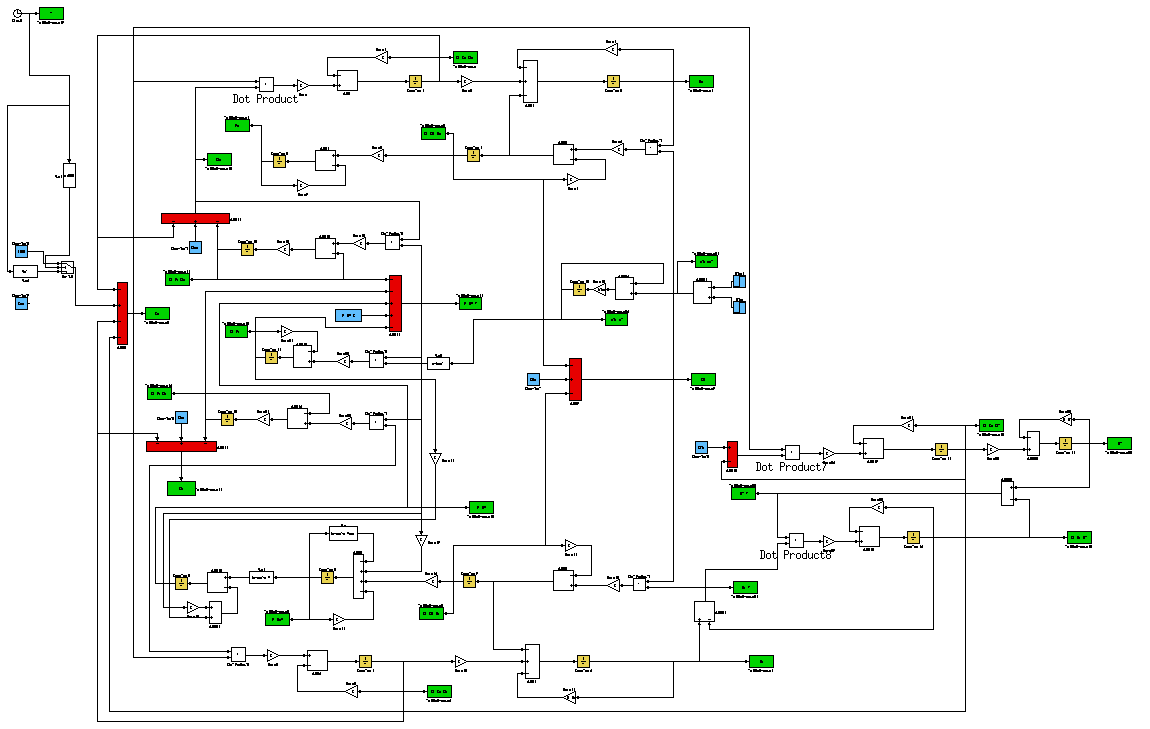
| - | + | To characterize the T-REX device, we developed a [https://2009.igem.org/Team:Bologna/Modeling mathematical model] of the whole testing circuit considering the interaction between all its parts. Model equations were implemented in Simulink (Fig. 4) to simulate the circuit and it intermediate parts. After the identification of model parameters on the data obtained from experiments with the intermediate parts, the GFP level as a function of the TRANS/CIS affinity was predicted by simulating the whole testing circuit . | |
| - | + | [[Image:ModelSandro.png|center|900px|thumb|<center>Figure 4 - Simulink Model</center>]] | |
| - | + | ||
<br><br> | <br><br> | ||
| - | <font face="Calibri" font size="5" color="#000000"><b> | + | <font face="Calibri" font size="5" color="#000000"><b>Characterization of Parts</b></font> |
<br><br> | <br><br> | ||
| - | + | The Testing Circuit is constituted by intermediate parts that were experimentally characterized to obtain date for the model paramiter identification. Details of this characterization are available in the [https://2009.igem.org/Team:Bologna/Wetlab wet-lab] section. | |
| - | + | ||
<br><br> | <br><br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
Latest revision as of 02:56, 22 October 2009
| HOME | TEAM | PROJECT | SOFTWARE | MODELING | WET LAB | PARTS | HUMAN PRACTICE | JUDGING CRITERIA |
|---|
(Trans-Repressor of Expression)
The aim of our project is the design of a standard device to control the synthesis of any protein of interest. This "general-purpose" device, implemented in E. coli, acts at the translational level to allow silencing of protein expression faster than using regulated promoters. We named this device T-REX (Trans Repressor of Expression).
T-REX consists of two new BioBricks:
- CIS-repressing, to be assembled upstream of the target protein coding sequence. It contains a ribosomal binding site (RBS);
- TRANS-repressor, complementary to the CIS-repressing and placed under the control of a different promoter. For a better repressive effectiveness, the TRANS sequence contains also a RBS cover, released in two versions of different length (either 4 or 7 nucleotides).
The longer version covers also 3 nucleotides of the Shine-Dalgarno sequence.
Transcription of the target gene yields a mRNA strand - containing the CIS-repressing sequence at its 5' end - available for translation into protein by ribosomes (see Fig. 1, left panel). When the promoter controlling the TRANS coding sequence is active, it drives the transcription of an oligoribonucleotide complementary to the CIS mRNA sequence. The TRANS/CIS RNA duplex prevents ribosomes from binding to RBS on target mRNA, thus silencing protein synthesis. The amount of the TRANS-repressor regulates the rate of translation of the target mRNA (see Fig. 1, right panel)
CIS and TRANS Parts Design
To identify CIS-repressing and TRANS-repressor DNA sequences, we developed BASER software. We used it to seek for two complementary 50bp non-coding sequences, whose transcribed RNAs:
a) feature maximal free energy in the secondary structure (i.e. reducing the probability of its intra-molecular annealing);
b) have minimal unwanted interactions with genomic mRNA;
c) present a minimal probability of partial/shifted hybridization with complementary strands.
Here below are the CIS-repressing and TRANS-repressor sequences. More details about BASER and its functioning can be found in the software section.
| CIS-repressing | |||
| Prefix | non-coding TRANS target | RBS | Suffix |
| GAATTCGCGGCCGCTTCTAGAG | AACACAAACTATCACTTTAACAACACATTACATATACATTAAAATATTAC | AAAGAGGAGAAA | TACTAGTAGCGGCCGCTGCAG |
| TRANS-repressor (4) | |||
| Prefix | RBS cover | non-coding TRANS | Suffix |
| GAATTCGCGGCCGCTTCTAGAG | CTTT | GTAATATTTTAATGTATATGTAATGTGTTGTTAAAGTGATAGTTTGTGTT | TACTAGTAGCGGCCGCTGCAG |
| TRANS-repressor (7) | |||
| Prefix | RBS cover | non-coding TRANS | Suffix |
| GAATTCGCGGCCGCTTCTAGAG | CCTCTTT | GTAATATTTTAATGTATATGTAATGTGTTGTTAAAGTGATAGTTTGTGTT | TACTAGTAGCGGCCGCTGCAG |
Testing Circuit
In order to test our T-REX device, we developed the following genetic circuit (Fig. 2):
The CIS-repressing sequence is assembled upstream of lac I (BBa_C0012), therefore the synthesis of LacI should be silenced/damped by the constitutively transcribed TRANS-repressor mRNA. To detect silencing of LacI, due to the action of T-REX, we realized a new inverter (BBa_K201001) consisting of a promoter regulated by LacI (BBa_K201008) and a GFP reporter (BBa_J04031).
We expect that a TRANS-repressor oligoribonucleotide with high affinity to CIS-repressing mRNA, inhibits the translation of LacI and then determines a maximally expressed GFP. Otherwise, in case of low TRANS/CIS affinity one should expect partially (or completely) repressed GFP expression.
To maximize the probability to silence the CIS transcript and switch on the GFP, we decided to use a high copy number (HCN) plasmid (pSB1A2) for the TRANS-repressor and a low copy number (LCN) plasmid (pSB3K3) for the LacI generator.
If the GFP inverter is unable to reveal the LacI reduction due to T-REX action, because of a high level of the free LacI concentration, IPTG can be supplied to reduce free LacI. In fact, the sensitivity of the GFP inverter to LacI variations depends on free LacI concentration. Using IPTG is thus possible to set actual LacI value in the region where the inverter has the highest sensitivity.
Mathematical Model
To characterize the T-REX device, we developed a mathematical model of the whole testing circuit considering the interaction between all its parts. Model equations were implemented in Simulink (Fig. 4) to simulate the circuit and it intermediate parts. After the identification of model parameters on the data obtained from experiments with the intermediate parts, the GFP level as a function of the TRANS/CIS affinity was predicted by simulating the whole testing circuit .
Characterization of Parts
The Testing Circuit is constituted by intermediate parts that were experimentally characterized to obtain date for the model paramiter identification. Details of this characterization are available in the wet-lab section.
 "
"