Team:Bologna/Project
From 2009.igem.org
| HOME | TEAM | PROJECT | SOFTWARE | MODELING | WET LAB | PARTS | HUMAN PRACTICE | JUDGING CRITERIA |
|---|
(Trans-Repressor of Expression)
The aim of our project is the design of a standard device to control the synthesis of any protein of interest. This "general-purpose" device, implemented in E. coli, acts at the translational level to allow silencing of protein expression faster than using regulated promoters. We named this device T-REX (Trans Repressor of Expression).
T-REX consists of two new BioBricks:
- CIS-repressing, to be assembled upstream of the target protein coding sequence. It contains a ribosomal binding site (RBS);
- TRANS-repressor, complementary to the CIS-repressing and placed under the control of a different promoter. For a better repressive effectiveness, the TRANS sequence contains also a RBS cover, released in two versions of different length (either 4 or 7 nucleotides).
The longer version covers also 3 nucleotides of the Shine-Dalgarno sequence.
Transcription of the target gene yields a mRNA strand - containing the CIS-repressing sequence at its 5' end - available for translation into protein by ribosomes (see Fig. 1, left panel). When the promoter controlling the TRANS coding sequence is active, it drives the transcription of an oligoribonucleotide complementary to the CIS mRNA sequence. The TRANS/CIS RNA duplex prevents ribosomes from binding to RBS on target mRNA, thus silencing protein synthesis. The amount of the TRANS-repressor regulates the rate of translation of the target mRNA (see Fig. 1, right panel)
To identify CIS-repressing and TRANS-repressor complementary parts, we developed BASER software. We used it to seek for two complementary 50bp non-coding sequences, whose transcribed RNAs:
a) feature maximal free energy in the secondary structure (i.e. reducing the probability of its intra-molecular annealing);
b) have minimal unwanted interactions with genomic mRNA;
c) present a minimal probability of partial/shifted hybridization with complementary strands.
Here below are the CIS-repressing and TRANS-repressor sequences:
| CIS-repressing | |||
| Prefix | non-coding TRANS target | RBS | Suffix |
| GAATTCGCGGCCGCTTCTAGAG | AACACAAACTATCACTTTAACAACACATTACATATACATTAAAATATTAC | AAAGAGGAGAAA | TACTAGTAGCGGCCGCTGCAG |
| TRANS-repressor (4) | |||
| Prefix | RBS cover | non-coding TRANS | Suffix |
| GAATTCGCGGCCGCTTCTAGAG | CTTT | GTAATATTTTAATGTATATGTAATGTGTTGTTAAAGTGATAGTTTGTGTT | TACTAGTAGCGGCCGCTGCAG |
| TRANS-repressor (7) | |||
| Prefix | RBS cover | non-coding TRANS | Suffix |
| GAATTCGCGGCCGCTTCTAGAG | CCTCTTT | GTAATATTTTAATGTATATGTAATGTGTTGTTAAAGTGATAGTTTGTGTT | TACTAGTAGCGGCCGCTGCAG |
More details about BASER and its functioning can be found in the software section.
The Genetic Circuits
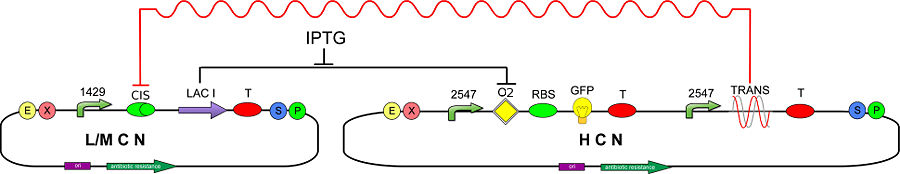
In order to test and characterize our T-REX device, we developed the following genetic circuit (Fig 2):
The CIS-repressing sequence is assembled upstream of LacI (BBa_C0012), therefore the synthesis of LacI should be silenced/damped by the constitutively transcribed TRANS-repressor mRNA. To detect silencing of LacI, due to the action of T-REX, we realized a new inverter (BBa_K201001) consisting of a promoter regulated by LacI (BBa_K201008) and a GFP reporter (BBa_J04031).
We expect that a TRANS-repressor oligoribonucleotide with high affinity to CIS-repressing mRNA, inhibits the translation of LacI and then determines a maximally expressed GFP. Otherwise, in case of low TRANS/CIS affinity one should expect partially (or completely) repressed GFP expression.
To maximize the probability to silence the CIS transcript and switch on the GFP, we decided to use a high copy number (HCN) plasmid (pSB1A2) for the TRANS-repressor and a low copy number (LCN) plasmid (pSB3K3) for the LacI generator.
If the GFP inverter is unable to reveal the LacI reduction due to T-REX action, because of a high level of the free LacI concentration, IPTG can be supply to reduce free LacI. In fact, the sensitivity of the GFP inverter to LacI variations depends on free LacI concentration. Using IPTG is thus possible to set actual LacI value in the region where the inverter has the highest sensitivity.
The circuit above represents the ON configuration of our T-REX device: TRANS-repressor is constitutively expressed and CIS-repressing is assembled upstream the LacI gene (BBa_C0012), while the reporter protein GFP (BBa_J04031) is assembled under the control of another promoter, regulated by LacI natural operator O2.
CIS-repressing and TRANS-repressor mRNAs bind together, preventing LacI translation and allowing GFP production.
Using some IPTG, the repression action of T-REX become easily, because the concentration of free LacI in the cells is reduced.
To prove T-REX repression, GFP production level must be compared with the level produced by the OFF configuration of our T-REX device, that is the same circuit in absence of TRANS-repressor:
In this second case, LacI is regularly translated and, binding with its O2 natural operator, it blocks GFP production.
We choose to use a low copy number plasmid and a weak promoter (J23118) for the CIS-repressing/LacI production, and a high copy number plasmid and a strong promoter (J23100) for the TRANS-repressing, to make reporter production's variations easily to be detected.
Before realizing the whole T-REX device, we decided to analyze some preliminary circuits, in order to characterize every single BioBrick involved in it.
pSB1A2 vs pSB3K3
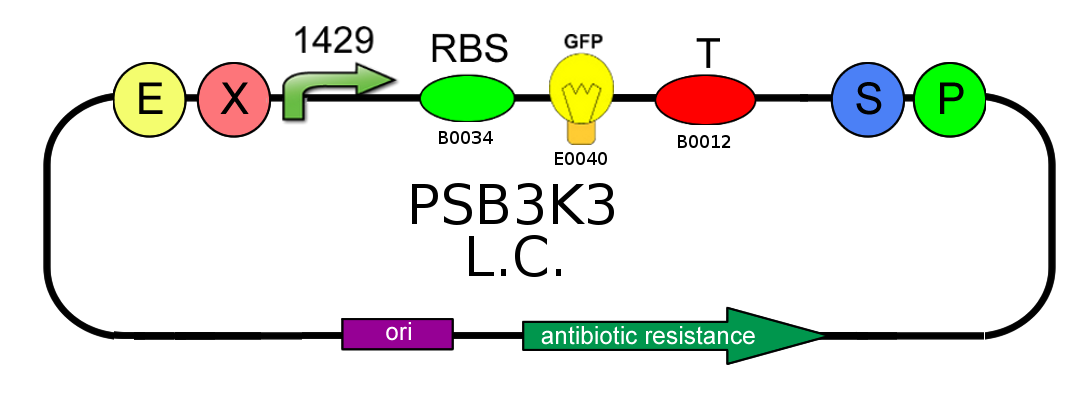
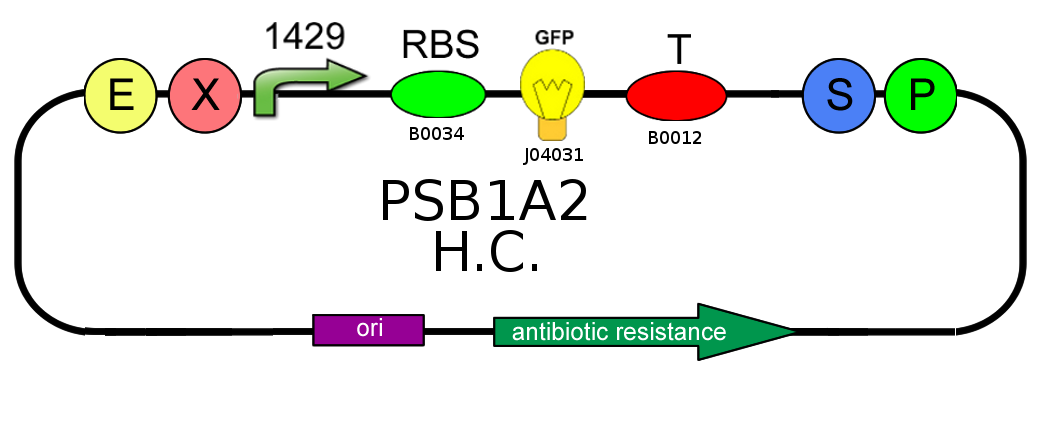
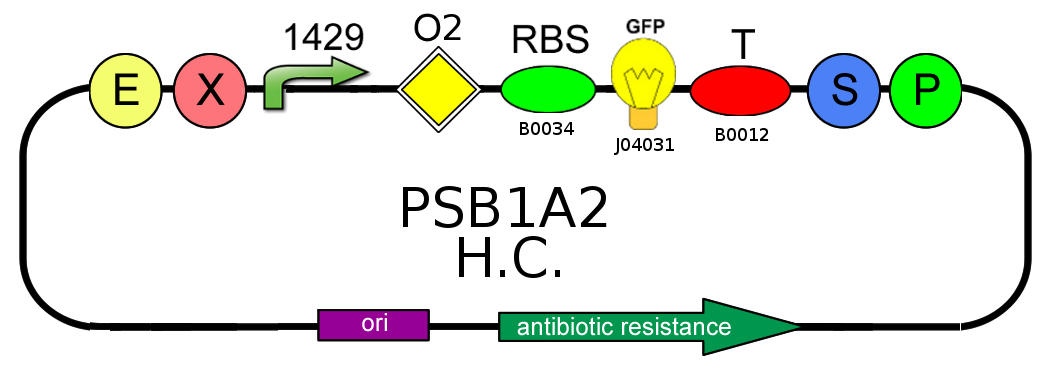
- In order to characterize the difference between the high copy number pSB1A2 and the low to medium copy number pSB3K3 plasmids, we analyzed production of BBa_I13504 (wild type GFP) under BBa_J23118 promoter (1429):
BBa_J23100 vs BBa_J23118
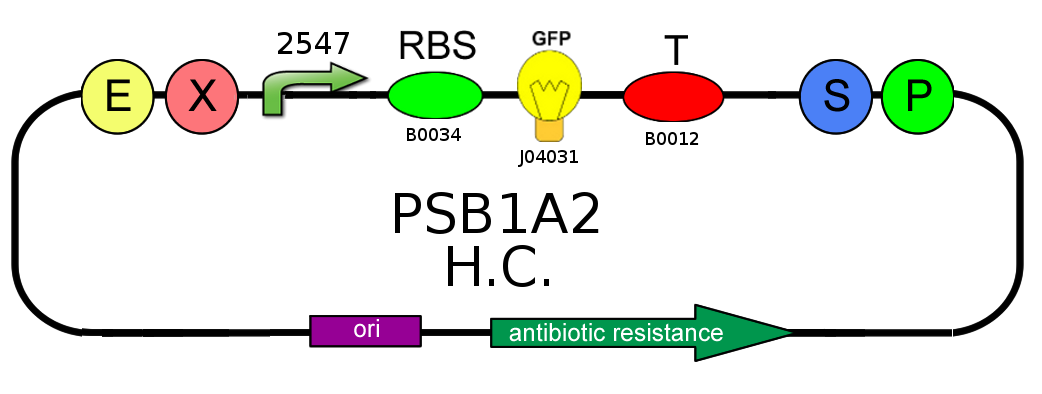
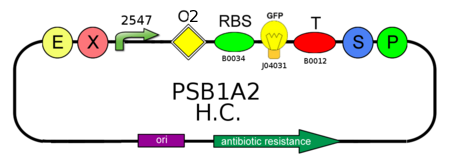
- In order to characterize the ratio between BBa_J23100 (strength 2547) and BBa_J23118 (strength 1429) promoters, we analyzed GFP (BBa_J04031) production on pSB1A2:
Presence vs Absence of LacI natural operator O2
- We needed to confirm that LacI natural operator O2 don't influence GFP production when LacI repressor is not present. We evaluate this on pSB1A2, using BBa_J04031 under the control of both BBa_J23118 and BBa_J23100, in presence and in absence of BBa_K079019:
Interaction of LacI repressor with its natural operator O2
- We studied interactions between LacI repressor and its natural operator O2, analyzing this two genetic circuits:
In the first circuit, we used different IPTG concentration in order to evaluate LacI repression strength. In the latter, we verified that LacI doesn't interfere with GFP production if its natural operator O2 is not present.
 "
"