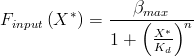Team:Cambridge/Notebook/Week5
From 2009.igem.org
(→Amplification) |
(→Amplification) |
||
| (47 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
{{Template:CambridgeNotepad}} | {{Template:CambridgeNotepad}} | ||
| + | |||
| + | ''The plate-reader data for the promoter/amplifier constructs is analysed - DNA-purification debugged - modelling the amplifier/theshold system - melA primers recieved - vio gene planned using GeneDesigner'' | ||
== Monday == | == Monday == | ||
| Line 14: | Line 16: | ||
[[Image:New_vio_plan.PNG |750px]] | [[Image:New_vio_plan.PNG |750px]] | ||
| + | |||
| + | ====Data Analysis==== | ||
| + | |||
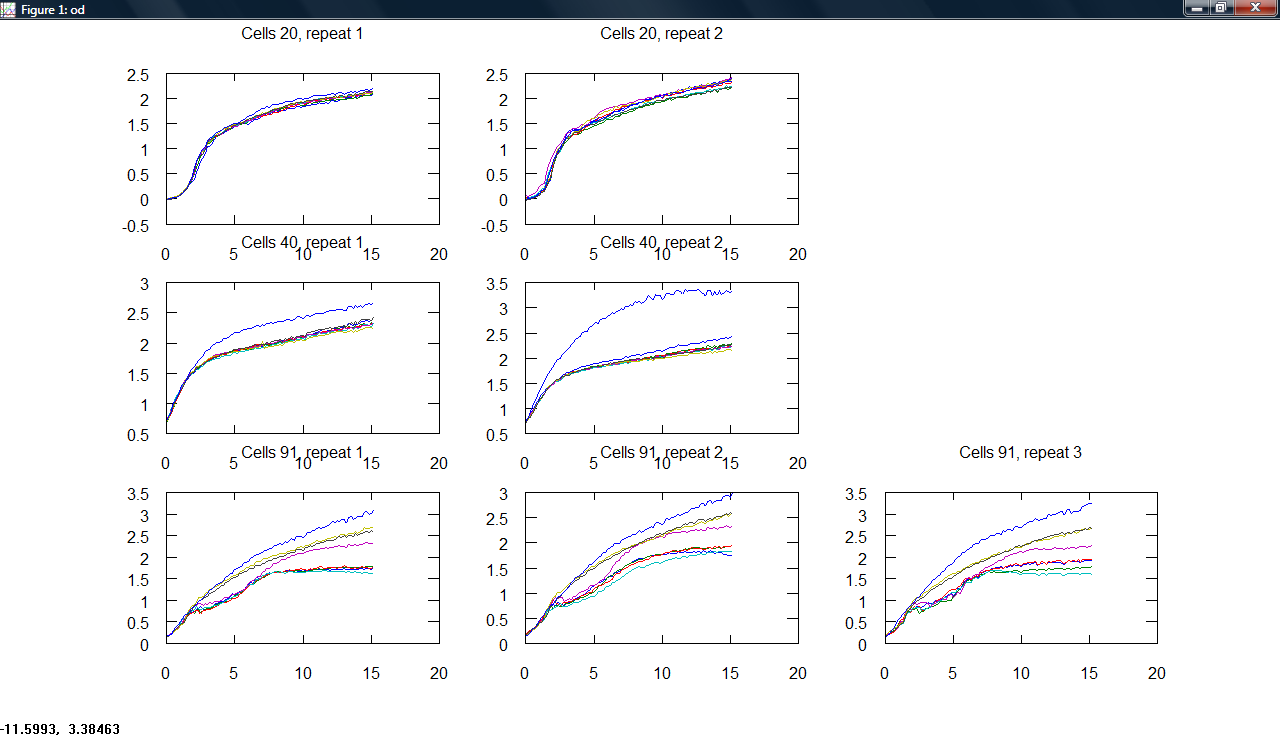
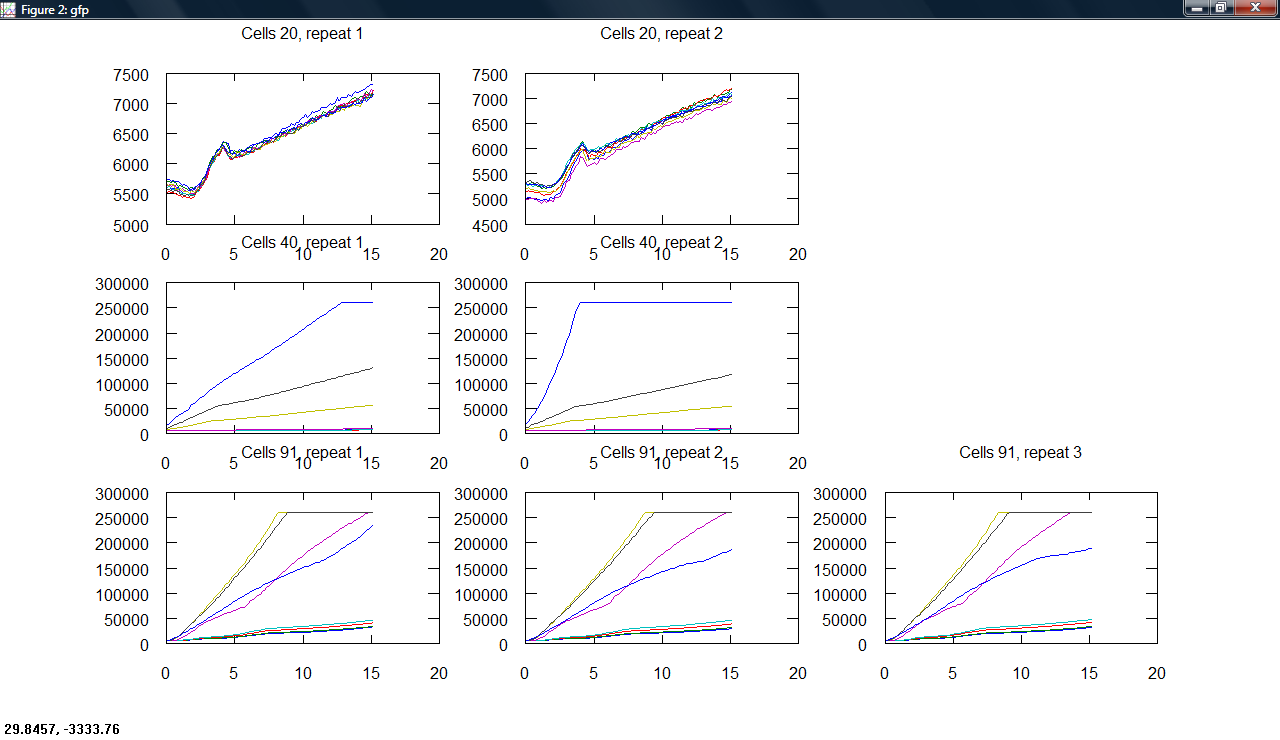
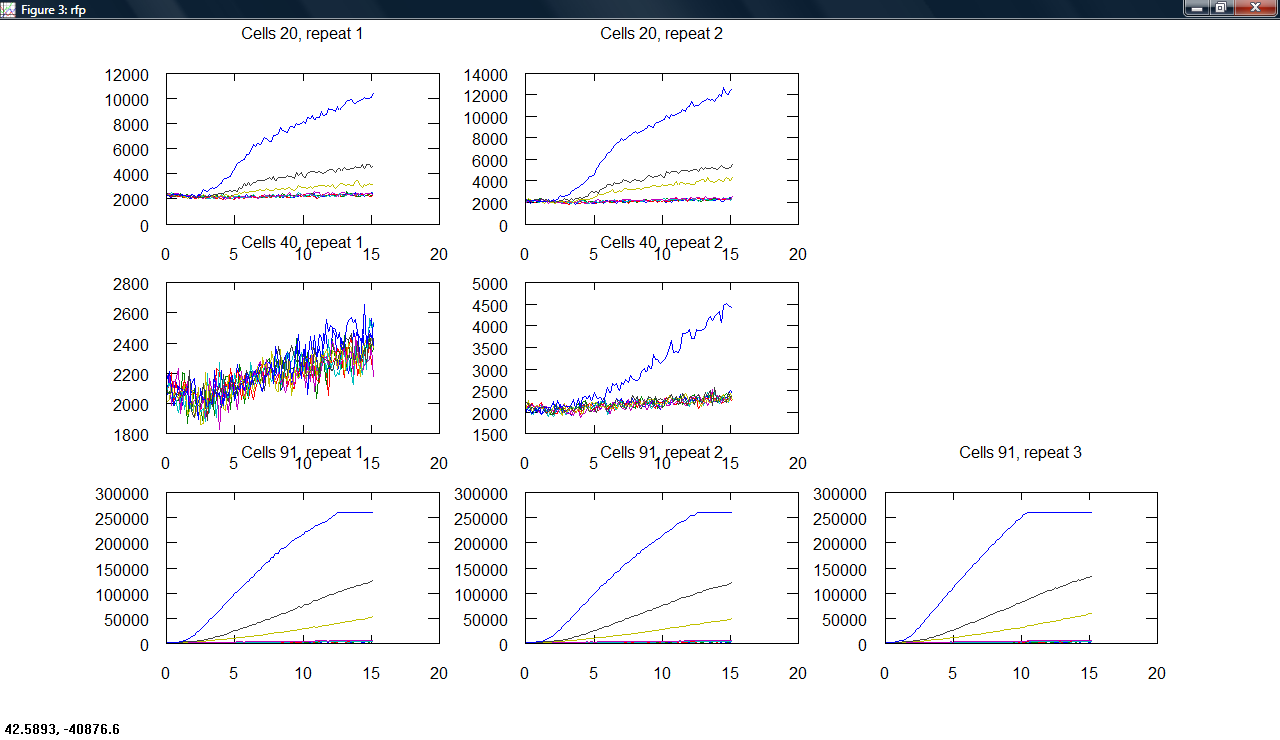
| + | Produced a plot making program, producing subplots across cell type and repeats for all measurements. | ||
| + | |||
| + | [[Image:Cambridge_Odgraphs.png|300px]][[Image:Cambridge_Gfpgraphs.png|300px]][[Image:Cambridge_Rfpgraphs.png|300px]] | ||
===Wet Work=== | ===Wet Work=== | ||
| Line 19: | Line 27: | ||
====Protocol de-bugging==== | ====Protocol de-bugging==== | ||
| - | For the | + | For the threshold device project, we need to amplify pSB3K3 as a low-copy number plasmid vector in order to transfer the activator constructs into low copy number plasmid. We tried 4 different PCR reaction mixtures, and used a Zymogen PCR wash kit before running them on an 0.8% Agarose E-gel. After looking at the gel, the ladder was quite smeared and no PCR product was visible. It was found that the samples run on the gel had a very low DNA concentration. However, some PCR solution was left before the wash kit was used, and using the Nano-Drop it was determined that it had a high concentration of DNA, so the PCR worked, we just need to refine the protocols after PCR. |
| - | ==== | + | ====Threshold Device==== |
Mini-prepped I746374, I746375, I746380, and I746391; incubated overnight cultures of the rest of the 11 activator constructs in preparation for miniprep. | Mini-prepped I746374, I746375, I746380, and I746391; incubated overnight cultures of the rest of the 11 activator constructs in preparation for miniprep. | ||
| Line 78: | Line 86: | ||
[[Image:Vio_E.PNG]] | [[Image:Vio_E.PNG]] | ||
| + | |||
| + | The protein structures given for each gene were also compared with vio proteins from other organisms, ( ''Janthinobacterium lividum'' and Duganella sp. B2) to confirm that the ORFs were coding for the correct protein. | ||
| + | |||
| + | ====Modelling==== | ||
| + | |||
| + | Using our model for the rate of transcription from pBAD, dependent on arabinose concentration: | ||
| + | |||
| + | [[Image:Cambridge Eq1.gif]] , where [[Image:Cambridge Eq2.gif]] | ||
| + | |||
| + | we discussed how this model would behave, and how to determine the parameters. | ||
| + | |||
| + | The behaviour of this model to changing arabinose concentrations is shown in these graphs | ||
| + | |||
| + | [[Image:Cambridge_Repressor_arabinose.png|500px]] | ||
| + | |||
| + | [[Image:Cambridge_Transcription_arabinose.png|500px]] | ||
===Wet Work=== | ===Wet Work=== | ||
| - | ==== | + | ==== Threshold Device ==== |
| - | Continued to refine protocols for successful PCR and purification of pSB3K3. | + | Continued to refine protocols for successful PCR and purification of pSB3K3; tried 4 different PCR mixtures, all of which were successful. |
Miniprepped all remaining amplifier constructs. | Miniprepped all remaining amplifier constructs. | ||
| Line 91: | Line 115: | ||
Yesterdays transformations were successful, continued to incubate to trigger pigment production. | Yesterdays transformations were successful, continued to incubate to trigger pigment production. | ||
| - | |||
| - | + | {{Template:CambridgeNewPage}} | |
| - | + | ||
== Wednesday == | == Wednesday == | ||
| Line 107: | Line 129: | ||
Removed restriction sites from the new vio plan (shown on yesterdays page) and send it to Jim, along with the ApE plasmid editor file of the operon. After getting the go-ahead from Jim, this plan will be send to DNA2.0 for synthesis. | Removed restriction sites from the new vio plan (shown on yesterdays page) and send it to Jim, along with the ApE plasmid editor file of the operon. After getting the go-ahead from Jim, this plan will be send to DNA2.0 for synthesis. | ||
| - | + | Drafted the design criteria for Violacein synthesis: | |
| + | :*The gene consists of five open reading frames (vioA-E) each preceded by a ribosome binding site (part B0034). The whole sequence is held within the BioBrick registry prefix and suffix | ||
| + | :*The DNA sequence of the prefix and suffix CAN NOT be altered in any way | ||
| + | :*Ideally the operon would be under the control of the repressible promoter suggested by Jeremy. This promoter would be upstream of the operon and ribosome binding sites, but downstream of the prefix | ||
| + | :*With the exception of the prefix and suffix (which cannot be altered) the following restriction sites are forbidden within the operon: EcoRI, XbaI, SpeI, PstI, NotI | ||
| + | :*If possible, we would like the genes to be optimised for E. coli and preferably B. subtilis as well | ||
| + | :*In order to easy gene separation, we would like restriction sites on either side of the vio C and the vio D genes. | ||
| + | |||
| + | ====Modelling==== | ||
| + | |||
| + | We have now developed several versions of our model for the amplifier. These involve a simple model, positive feedback and degradation of the activator protein. We also included dependence on a parameter for input transcription, describing changes in both arabinose concentration and binding properties. | ||
| + | |||
| + | We have some interesting results for the positive feedback latch. Our ''continuous'' model indicates that whatever the transcriptional input, the reporter is produced in the same way. This means that the latch would be switched at any input level. We plan to look at stochastic modelling to see if the same applies here. | ||
| + | |||
| + | We produced several graphs to show what happens when different parameters are changed for the latch. The most important is the following, showing the very small effect of changing transcriptional input - encapsulating the effect of different arabinose concentrations. | ||
| + | |||
| + | [[Image:Cambridge_Latch_changing_Cin.png|600px]] | ||
| + | |||
| + | ====Data Analysis==== | ||
| + | |||
| + | We now have fluorescence rate calculated for all runs, and have normalised this for od. The next step is to take the steady state fluorescence rate per cell, and plot this against arabinose concentration. This should allow us to show the effect of the amplifier construct. | ||
===Wet Work=== | ===Wet Work=== | ||
| Line 119: | Line 161: | ||
Did a PCR to amplify the melanin fragment that will be made into a BioBrick. Purpose was to measure the length of the MelA fragment, check that the A and F primers were functional (as they are used in every PCR cycle in the plan), and produce samples for testing our different DNA clean-up protocols. We used the Finnzymes MasterMix with added DMSO and ran on an ethidium bromide gel. | Did a PCR to amplify the melanin fragment that will be made into a BioBrick. Purpose was to measure the length of the MelA fragment, check that the A and F primers were functional (as they are used in every PCR cycle in the plan), and produce samples for testing our different DNA clean-up protocols. We used the Finnzymes MasterMix with added DMSO and ran on an ethidium bromide gel. | ||
| - | (gel | + | [[Image:Gel.jpg | 400px]] |
| + | |||
| + | Gel was run at 80V for 45mins. The two samples were made of the same master mix (as in Finnzymes PCR protocol) with added 1.5ul of DMSO to each 50ul sample. The bands are approximately 2kb, which matches the 1.9kb size of the melA fragment. The smaller bands near the bottom of the gel are the primers. | ||
| + | |||
| + | ====Threshold Device==== | ||
| + | |||
| + | Made glycerol stocks for -20 freezer of all activator constructs. | ||
| + | |||
| + | Continued to de-bug DNA clean-up after PCR, PCR amplified more pSB3K3. | ||
| + | |||
| + | {{Template:CambridgeNewPage}} | ||
| + | |||
| + | ==Thursday== | ||
| + | |||
| + | ===Dry Work=== | ||
| + | |||
| + | ====Data Analysis==== | ||
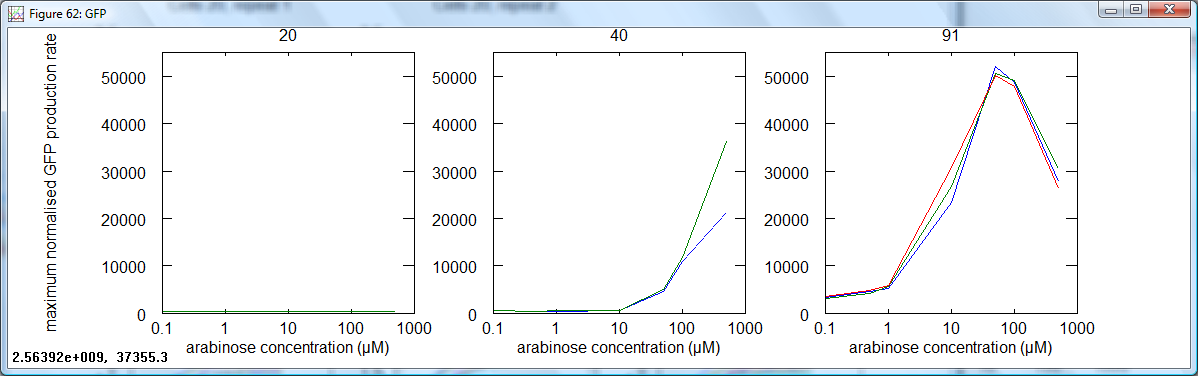
| + | *For each data set, the maximum normalised fluorescence rate was taken. This is the measurement used to calculate Relative Promoter Units, and by extention, RNA Polymerase per second (PoPS). These data were plotted for each cell type against concentration of arabinose: | ||
| + | |||
| + | [[Image:Cambridge_Maxraterfp.png|600px]] | ||
| + | |||
| + | [[Image:Cambridge_Maxrategfp.png|600px]] | ||
| + | |||
| + | The most important graphs above are those for GFP in cells 40 (pBAD) and 91 (amplifier). These are effectively the input and output for the amplifier device. One can see that the pBAD curve has no real increase until at least 10μM, whereas an equivalent large increase for the amplifier starts at about 1μM. Very low level transcription can be seen to be amplified. The amplified graph (91) starts to drop off at high concentrations: this may be due to toxicity. | ||
| + | |||
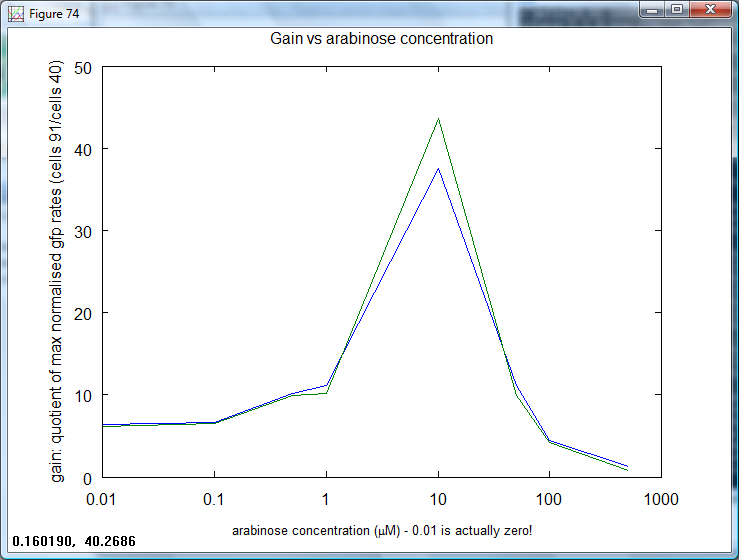
| + | *Next, in order to remove the effect of pBAD, the gain (ratio output/input) at each concentration was calculated: | ||
| + | |||
| + | [[Image:Cambridge_Gain_concentration.png|600px]] | ||
| + | |||
| + | This graph shows that the amplifier has steady gain at low concentrations. The point at "0.01" is actually that for zero, showing the basal amplification. What is interesting in this graph is that the gain drops off, well before the reduction of expression for cells 91. This is because cells 40 start to rise here. | ||
| + | |||
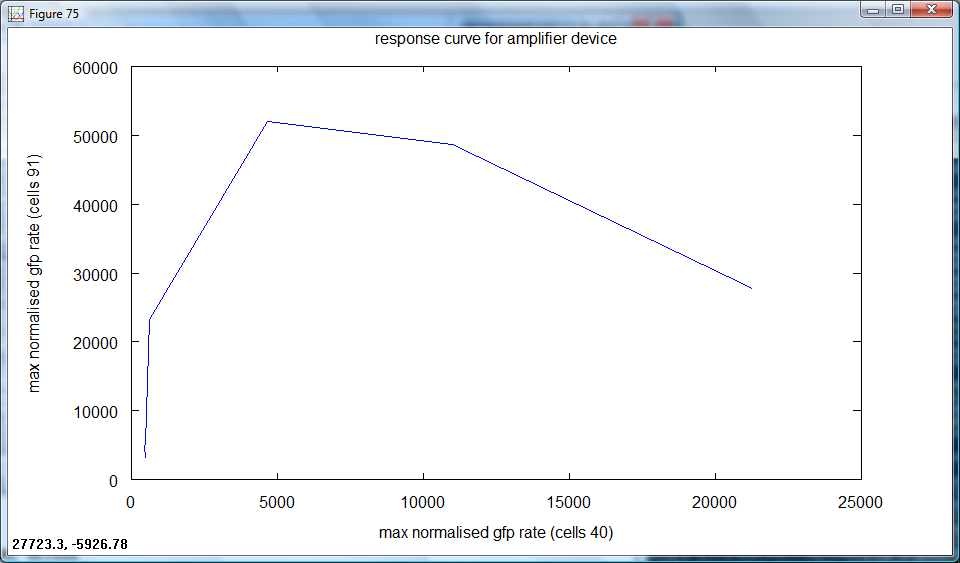
| + | *Finally, in order to fully abstract the behaviour of the amplifier device from arabinose concentration, the output signal was plotted directly against the input signal: | ||
| + | |||
| + | [[Image:Cambridge_Response_curve.png|600px]] | ||
| + | |||
| + | A linear device would have a straight line here. Instead we observe a rapid increase in output at a low range of input, followed by a plateau, then a decrease at higher input. There is scope for the device to be used in both discrete and continuous applications. | ||
| + | |||
| + | === Minutes from Lab Meeting === | ||
| + | |||
| + | *Melanin and Violacein | ||
| + | :*Characterisation of the pigments - consider LCMS | ||
| + | :*Plate all colours as streaks on the same plate, for presentation purposes | ||
| + | :*Check the Melanin rbs is compatible with B. subtilis | ||
| + | :*We could probably have a think about why our pigments don't grow in liquid culture. | ||
| + | |||
| + | *Carotenoids | ||
| + | :*Check the gene homology for the different genes | ||
| + | :*Contact Edinburgh to ask about the promoter | ||
| + | :*Double-check the orange colour is actually being formed by the genes we think they are | ||
| + | :*The Roche strain can only be used for testing, not for the actual iGEM product | ||
| + | |||
| + | *DNA purification | ||
| + | :*Overall, we are still not sure why the column washes are not working | ||
| + | :*Suggested that we try purification with 0.5M Ammonium acetate | ||
| + | |||
| + | *Melanin retention | ||
| + | :*More controls needed to check whether the melanin diffusion out of the cell is the cause of the staining. | ||
| + | |||
| + | *Modelling | ||
| + | :*Presented the continuous modelled behaviour of the latch: to a large extent independent of input transcription. This means it is likely to turn on under basal input. | ||
| + | :*Stochastic modelling may agree or disagree with this finding. | ||
| + | |||
| + | *Overall Plan | ||
| + | :*We need a lab meeting to set out the following: | ||
| + | ::*What we want | ||
| + | ::*What is feasible | ||
| + | ::*What is justifiable within the overall project plan | ||
| + | :*Work towards the end-result, and the talk/presentation at MIT | ||
| + | ::*Results with theoretical underpinning | ||
| + | ::*What we have managed and what our model days we could achieve | ||
| + | ::*Finished PRODUCT | ||
| + | :*Ideas for finished product - dipstick, 'lollypop' etc. | ||
| + | |||
| + | ====Modelling==== | ||
| + | *Basic promoter-activator-phage promoter system modelled. A variety of parameters changed and the effects collated. | ||
| + | *Addition of positive feedback for first 'latch' design - Note that the same output is gained for any input, so it would destroy any natural switching level on-off property of the activators. | ||
| + | *Began to develop 'threshold' switching level system as detailed at bottom of modelling page. Considered different mechanisms by which it could work: | ||
| + | |||
| + | :*activator and repressor bind to different sites on operator, both can be bound at once, but transcription only occurs when the activator alone is bound. | ||
| + | |||
| + | :*activator and repressor compete for binding to DNA; they do not necessarily bind the same site, but if one is present the other cannot bind. | ||
| + | |||
| + | :*the activator prevents production of the repressor by inhibiting transcription, but it is only capable of doing this once it has reached a particular concentration. This is effectively just the activator system with a second step, as the switching level depends on the properties of the promoter, not the quantity of repressor present. It would still be interesting to see what happens when this extra level is included and whether this helps remove basal transcription level problems. | ||
| + | |||
| + | ===Wet Work=== | ||
====Amplification==== | ====Amplification==== | ||
| Line 125: | Line 251: | ||
Overnight culture of a single colony of J69591, phenotypically confirmed, in preparation to make glycerol stocks, steak out on a plate, and miniprep. This will be the parent colony for all of our characterizations. | Overnight culture of a single colony of J69591, phenotypically confirmed, in preparation to make glycerol stocks, steak out on a plate, and miniprep. This will be the parent colony for all of our characterizations. | ||
| - | + | ====Melanin Retention==== | |
| + | |||
| + | Streaks of the strains requested from Philip Oliver containing multi-drug transporter knockouts arrived - MC1060 containg a deletion of the acrAB operon, and MG1655 containing a deletion of the acrF gene (part of the acrEF operon). Competent cell stocks were prepared in readiness for the next day's transformations with the MelA-containing plasmid. | ||
| + | |||
| + | {{Template:CambridgeNewPage}} | ||
| + | |||
| + | ==Friday== | ||
| + | |||
| + | ===Wet Work=== | ||
| + | |||
| + | ====Melanin Pigment==== | ||
| + | |||
| + | Made controls (as suggested in the meeting yesterday) to confirm that the melanin pigment was being produced by the bacteria with the MelA gene, rather than by oxidation of plate substrates. Schematic is shown below - all plates contain tyrosine, copper sulfate, and IPTG. Brown colour represents melanin bacteria, while pale stain represents the TOP10 control species. All plates were stored over the weekend at 37 degrees. | ||
| + | |||
| + | [[Image:Mel_controls.PNG]] | ||
| + | |||
| + | |||
| + | ====Melanin Retention==== | ||
| + | |||
| + | Electroporation was used to transform the MC1060 (containing an acrAB deletion) and MG1655 (containing an acrF deletion) strains with the MelA-containing plasmid before plating them out onto ampicillin plates. Hopefully these should show limited melanin diffusion into the agar medium and delineated black colonies after a spell in the incubator. | ||
| + | |||
| + | ====DNA Wash==== | ||
| + | |||
| + | As part of troubleshooting the column wash procedure, the QUIGEN kit was used to purify the DNA after a PCR. The gel below shoes, from left to right, 2-log ladder, unwashed PCR-product, 1/10 diluted PCR product, and washed DNA. The QUIGEN kit therefore produces a reasonable yield of pure DNA, and will be used in the melanin protocol which will be startedon tuesday. | ||
| + | |||
| + | (GEL PICTURE) | ||
| + | |||
| + | ===Dry Work=== | ||
| + | ====Modelling==== | ||
| + | The threshold switching system was modelled, giving model 6.1 (transcription only when activator bound) and model 6.2 (competition between activator and repressor). A plot of output against repressor present is given for a set input which shows the expected switching off of output at a threshold (this is the reverse of an activator switch on, effectively). | ||
| + | [[Image:Cambridge_model6_2.png | 750px ]] | ||
| + | |||
| + | ====Violacein Synthesis==== | ||
| + | |||
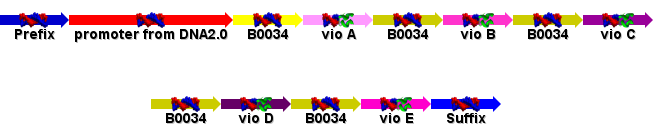
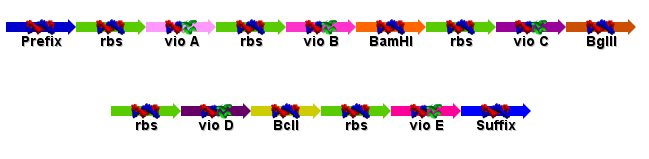
| + | Sent the order to DNA2.0! The GeneDesigner file was send, along with the ApE files for the vio operon and the vector we hope to splice the operon into (pPB3K3). The final design is shown below: | ||
| + | |||
| + | [[Image:Design_sent_to_DNA_2.0.PNG]] | ||
| + | |||
| + | Design specifications were also finalised: | ||
| + | :*The gene consists of five open reading frames (vioA-E) each preceded by a ribosome binding site. The whole sequence is held within the BioBrick registry prefix and suffix | ||
| + | :*The DNA sequence of the prefix and suffix cannot be altered in any way | ||
| + | :*As overexpression of the violacein operon can be toxic for the bacteria, we would like the operon under control of an inducible/repressible promoter, such as the T5 promoter suggested by Jeremy. | ||
| + | :*With the exception of the prefix and suffix (which cannot be altered) the following restriction sites are forbidden within the operon: EcoRI, XbaI, SpeI, PstI, NotI | ||
| + | :*If possible, we would like the genes to be optimised for E. coli and preferably B. subtilis as well | ||
| + | :*In order to easy gene separation, we would like restriction sites on either side of the vio C and the vio D genes. Ideally ones with compatible overhanging ends such as BamHI, BclI and BglII | ||
| + | |||
| + | Vector: pJexpress 401-406 (T5) Cloning Cassette | ||
| - | <!--Do not remove the first and last lines in this page!--> | + | <!--Do not remove the first and last lines in this page!-->{{Template:CambridgeBottom}} |
Latest revision as of 08:32, 3 September 2009
Categories :
Project :
-
Overview
Sensitivity Tuner
--- Characterisation
--- Modelling
Colour Generators
--- Carotenoids (Orange/Red)
--- Melanin (Brown)
--- Violacein (Purple/Green)
The Future
Safety
Notebook :
Team Logistics :
Week 5
The plate-reader data for the promoter/amplifier constructs is analysed - DNA-purification debugged - modelling the amplifier/theshold system - melA primers recieved - vio gene planned using GeneDesigner
Monday
Dry Work
Violacein Synthesis
Used GeneDesigner from DNA2.0 to plan what we wanted synthesised, taking into account the different genes in the operon. This would be synthesised with codon optimisation for E. coli. All forbidden restriction sites have been removed (EcoRI, XbaI, SpeI, PstI, NotI)
Data Analysis
Produced a plot making program, producing subplots across cell type and repeats for all measurements.
Wet Work
Protocol de-bugging
For the threshold device project, we need to amplify pSB3K3 as a low-copy number plasmid vector in order to transfer the activator constructs into low copy number plasmid. We tried 4 different PCR reaction mixtures, and used a Zymogen PCR wash kit before running them on an 0.8% Agarose E-gel. After looking at the gel, the ladder was quite smeared and no PCR product was visible. It was found that the samples run on the gel had a very low DNA concentration. However, some PCR solution was left before the wash kit was used, and using the Nano-Drop it was determined that it had a high concentration of DNA, so the PCR worked, we just need to refine the protocols after PCR.
Threshold Device
Mini-prepped I746374, I746375, I746380, and I746391; incubated overnight cultures of the rest of the 11 activator constructs in preparation for miniprep.
Pigments
Melanin
Retransformed melanin plasmid into Top10, and transformed it into MG1655 to (hopefully) achieve better expression. Plated on plates optimized for pigment production, and prepared for solid-phase pigment extraction
Violacein
Retransformed violacein plasmid into Top10, and transformed it into MG1655 to (hopefully) achieve better expression.
Mixing pigments
Plated Top10 transformed with melanin plasmid and violacein plasmid and the Roche strain transformed with Duncan’s orange plasmid on the same plate, optimized for melanin production (as this is the only pigment that requires media supplementation) but without any antibiotic resistance.
Progress on Carotenoids (Monday)
Today we carried out restriction digest of K118006/CrtB (as recipient plasmid vector) using SpeI and PstI (two cuts both within suffix), and of K118005/CrtI (as gene insert) using XbaI and PstI (one cut within prefix, the other cut within suffix).
Recall that the sizes are: K118006/CrtB: insert: 949bp, backbone: 2079bp, total: 3028bp. K118005/CrtI: insert: 1498bp, backbone: 2079bp, total: 3577bp.
Hence we expect one band of about 3000bp for K118006/CrtB, and two bands of about 1500bp and 2100bp for K118005/CrtI, after restriction digest with their respective enzymes.
The restriction digest products were run on gel, and the results were shown below:
Lane 1: Log-2 DNA ladder (NEB). Lanes 2 and 3: Restriction digest products of K118006/CrtB (less DNA loaded in lane 3). Lane 4: Undigested K118006/CrtB (negative control). Lane 5 empty. Lane 6: Restriction digest products of K118005/CrtI. Lane 7: Undigested K118005/CrtI (negative control).
This indicated that the restriction digest was successful.
We then carried out gel extraction on Lane 2 and Lane 6, ligation with T4 ligase, and electroporation into E. coli Top10 cells. Results of transformation will be ready for inspection tomorrow.
Tuesday
Dry Work
Violacein gene
Confirmed that correct protein sequence was present for each gene, by using MUSCLE to compare the gene sequence given by GeneDesigner to the sequence in the NCBI database. All protein sequences were identical.
Prepared the GeneDesigner file and the ApE file of the violacein operon to be sent for sequencing. Jim suggested that we refactor the DNA so that each individual ORF is preceeded by an RBS (the biobrick part B0034) and a constituitive promoter added. DNA2.0 kindly agreed to provide the promoter sequence for us, and add it in along with the operon. We also included the vio E gene after reading the Sanchez paper: http://www3.interscience.wiley.com/cgi-bin/fulltext/112732008/HTMLSTART?CRETRY=1&SRETRY=0
- New Vio Plan
The protein structures given for each gene were also compared with vio proteins from other organisms, ( Janthinobacterium lividum and Duganella sp. B2) to confirm that the ORFs were coding for the correct protein.
Modelling
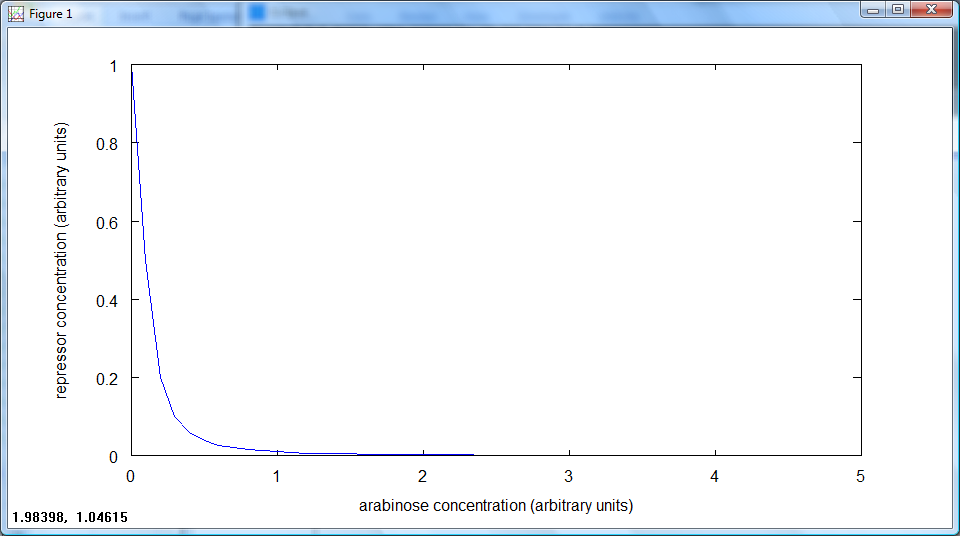
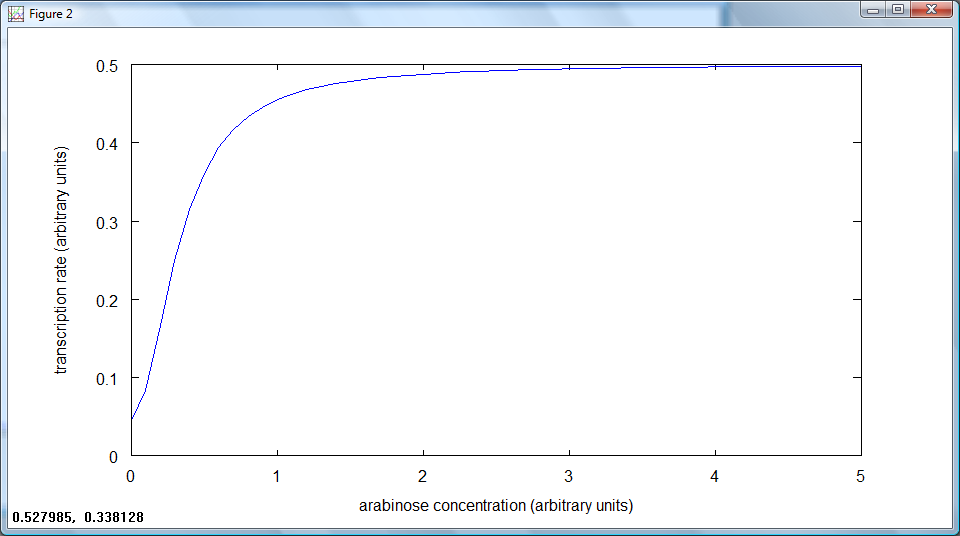
Using our model for the rate of transcription from pBAD, dependent on arabinose concentration:
we discussed how this model would behave, and how to determine the parameters.
The behaviour of this model to changing arabinose concentrations is shown in these graphs
Wet Work
Threshold Device
Continued to refine protocols for successful PCR and purification of pSB3K3; tried 4 different PCR mixtures, all of which were successful.
Miniprepped all remaining amplifier constructs.
Pigments
Yesterdays transformations were successful, continued to incubate to trigger pigment production.
Wednesday
Dry Work
Pseudomonas Green and Red
Received confirmation that we will be receiving clones containing the appropriate plasmids (YAY) soon.
Violacein
Removed restriction sites from the new vio plan (shown on yesterdays page) and send it to Jim, along with the ApE plasmid editor file of the operon. After getting the go-ahead from Jim, this plan will be send to DNA2.0 for synthesis. Drafted the design criteria for Violacein synthesis:
- The gene consists of five open reading frames (vioA-E) each preceded by a ribosome binding site (part B0034). The whole sequence is held within the BioBrick registry prefix and suffix
- The DNA sequence of the prefix and suffix CAN NOT be altered in any way
- Ideally the operon would be under the control of the repressible promoter suggested by Jeremy. This promoter would be upstream of the operon and ribosome binding sites, but downstream of the prefix
- With the exception of the prefix and suffix (which cannot be altered) the following restriction sites are forbidden within the operon: EcoRI, XbaI, SpeI, PstI, NotI
- If possible, we would like the genes to be optimised for E. coli and preferably B. subtilis as well
- In order to easy gene separation, we would like restriction sites on either side of the vio C and the vio D genes.
Modelling
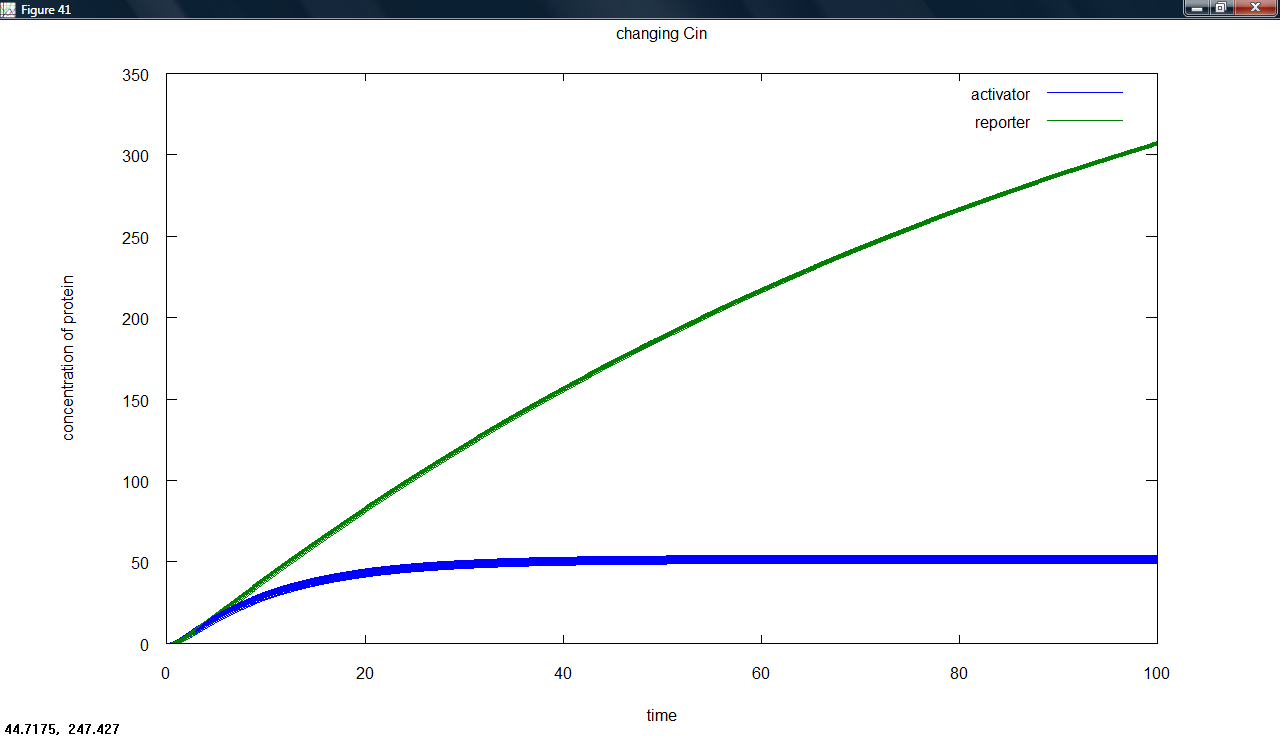
We have now developed several versions of our model for the amplifier. These involve a simple model, positive feedback and degradation of the activator protein. We also included dependence on a parameter for input transcription, describing changes in both arabinose concentration and binding properties.
We have some interesting results for the positive feedback latch. Our continuous model indicates that whatever the transcriptional input, the reporter is produced in the same way. This means that the latch would be switched at any input level. We plan to look at stochastic modelling to see if the same applies here.
We produced several graphs to show what happens when different parameters are changed for the latch. The most important is the following, showing the very small effect of changing transcriptional input - encapsulating the effect of different arabinose concentrations.
Data Analysis
We now have fluorescence rate calculated for all runs, and have normalised this for od. The next step is to take the steady state fluorescence rate per cell, and plot this against arabinose concentration. This should allow us to show the effect of the amplifier construct.
Wet Work
Pigment Stocks
Retransformed melanin and violacein pigment into MG1655 (previous attempt was unsuccessful)
Melanin Biobrick
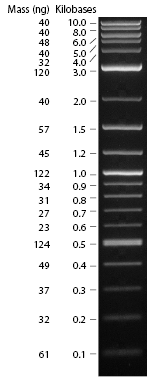
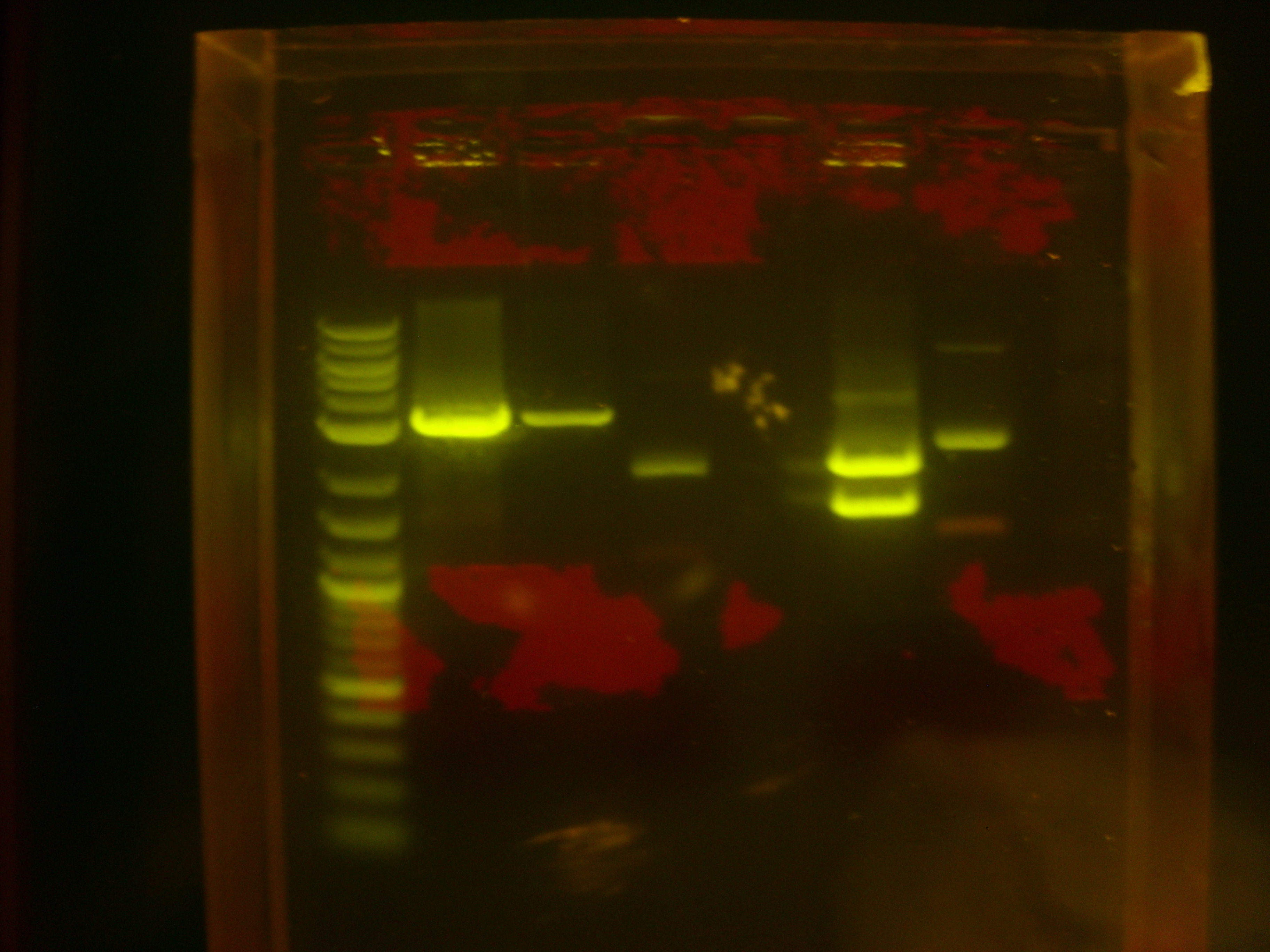
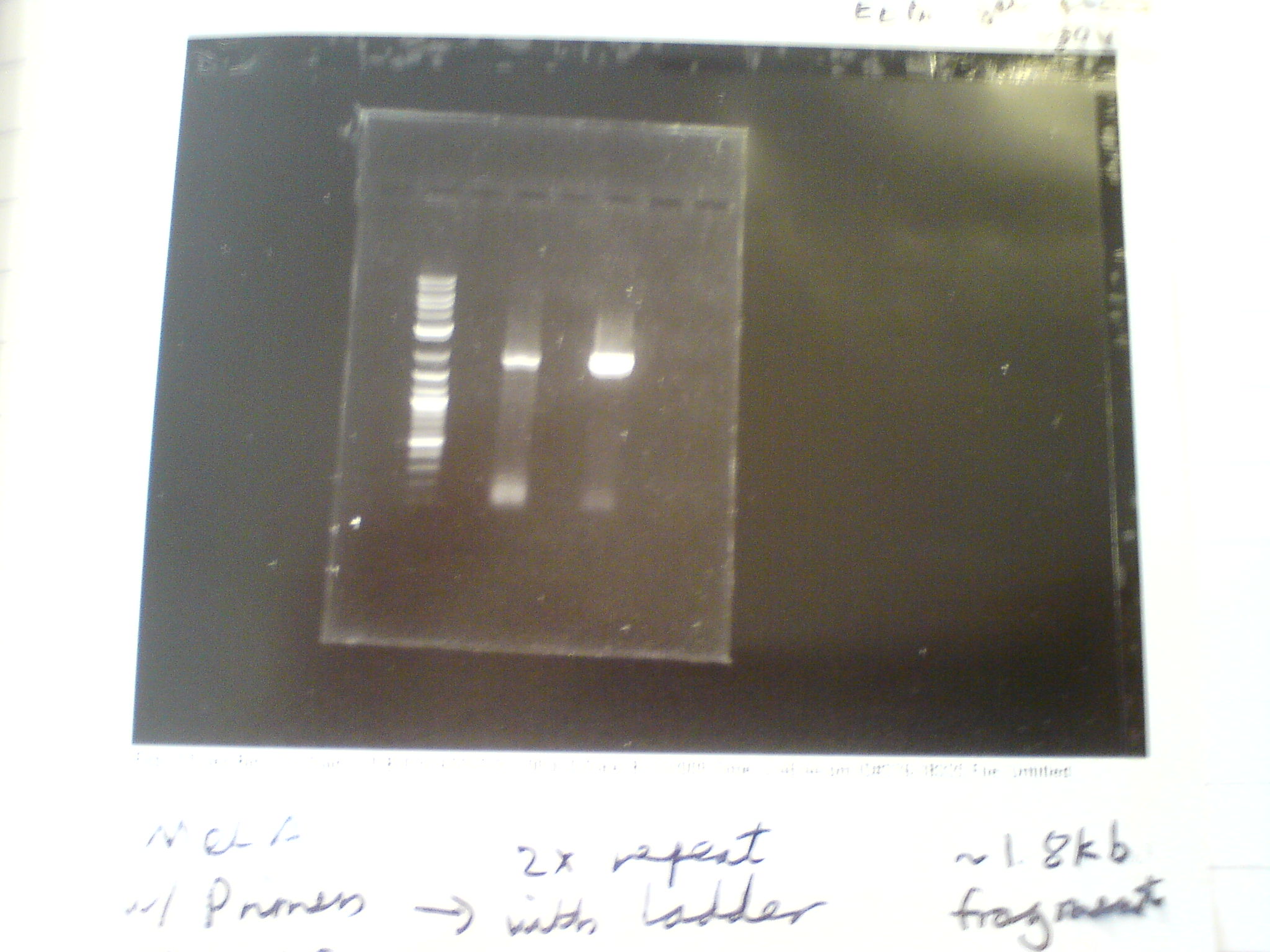
Did a PCR to amplify the melanin fragment that will be made into a BioBrick. Purpose was to measure the length of the MelA fragment, check that the A and F primers were functional (as they are used in every PCR cycle in the plan), and produce samples for testing our different DNA clean-up protocols. We used the Finnzymes MasterMix with added DMSO and ran on an ethidium bromide gel.
Gel was run at 80V for 45mins. The two samples were made of the same master mix (as in Finnzymes PCR protocol) with added 1.5ul of DMSO to each 50ul sample. The bands are approximately 2kb, which matches the 1.9kb size of the melA fragment. The smaller bands near the bottom of the gel are the primers.
Threshold Device
Made glycerol stocks for -20 freezer of all activator constructs.
Continued to de-bug DNA clean-up after PCR, PCR amplified more pSB3K3.
Thursday
Dry Work
Data Analysis
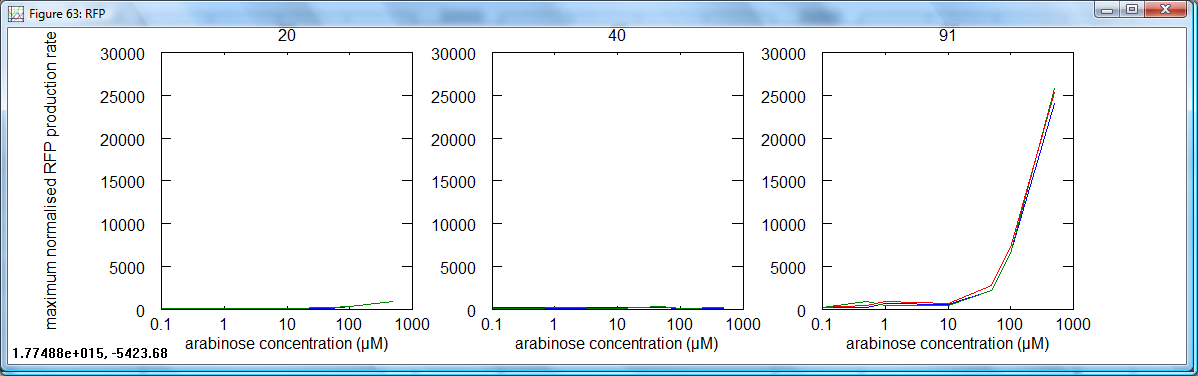
- For each data set, the maximum normalised fluorescence rate was taken. This is the measurement used to calculate Relative Promoter Units, and by extention, RNA Polymerase per second (PoPS). These data were plotted for each cell type against concentration of arabinose:
The most important graphs above are those for GFP in cells 40 (pBAD) and 91 (amplifier). These are effectively the input and output for the amplifier device. One can see that the pBAD curve has no real increase until at least 10μM, whereas an equivalent large increase for the amplifier starts at about 1μM. Very low level transcription can be seen to be amplified. The amplified graph (91) starts to drop off at high concentrations: this may be due to toxicity.
- Next, in order to remove the effect of pBAD, the gain (ratio output/input) at each concentration was calculated:
This graph shows that the amplifier has steady gain at low concentrations. The point at "0.01" is actually that for zero, showing the basal amplification. What is interesting in this graph is that the gain drops off, well before the reduction of expression for cells 91. This is because cells 40 start to rise here.
- Finally, in order to fully abstract the behaviour of the amplifier device from arabinose concentration, the output signal was plotted directly against the input signal:
A linear device would have a straight line here. Instead we observe a rapid increase in output at a low range of input, followed by a plateau, then a decrease at higher input. There is scope for the device to be used in both discrete and continuous applications.
Minutes from Lab Meeting
- Melanin and Violacein
- Characterisation of the pigments - consider LCMS
- Plate all colours as streaks on the same plate, for presentation purposes
- Check the Melanin rbs is compatible with B. subtilis
- We could probably have a think about why our pigments don't grow in liquid culture.
- Carotenoids
- Check the gene homology for the different genes
- Contact Edinburgh to ask about the promoter
- Double-check the orange colour is actually being formed by the genes we think they are
- The Roche strain can only be used for testing, not for the actual iGEM product
- DNA purification
- Overall, we are still not sure why the column washes are not working
- Suggested that we try purification with 0.5M Ammonium acetate
- Melanin retention
- More controls needed to check whether the melanin diffusion out of the cell is the cause of the staining.
- Modelling
- Presented the continuous modelled behaviour of the latch: to a large extent independent of input transcription. This means it is likely to turn on under basal input.
- Stochastic modelling may agree or disagree with this finding.
- Overall Plan
- We need a lab meeting to set out the following:
- What we want
- What is feasible
- What is justifiable within the overall project plan
- Work towards the end-result, and the talk/presentation at MIT
- Results with theoretical underpinning
- What we have managed and what our model days we could achieve
- Finished PRODUCT
- Ideas for finished product - dipstick, 'lollypop' etc.
Modelling
- Basic promoter-activator-phage promoter system modelled. A variety of parameters changed and the effects collated.
- Addition of positive feedback for first 'latch' design - Note that the same output is gained for any input, so it would destroy any natural switching level on-off property of the activators.
- Began to develop 'threshold' switching level system as detailed at bottom of modelling page. Considered different mechanisms by which it could work:
- activator and repressor bind to different sites on operator, both can be bound at once, but transcription only occurs when the activator alone is bound.
- activator and repressor compete for binding to DNA; they do not necessarily bind the same site, but if one is present the other cannot bind.
- the activator prevents production of the repressor by inhibiting transcription, but it is only capable of doing this once it has reached a particular concentration. This is effectively just the activator system with a second step, as the switching level depends on the properties of the promoter, not the quantity of repressor present. It would still be interesting to see what happens when this extra level is included and whether this helps remove basal transcription level problems.
Wet Work
Amplification
Overnight culture of a single colony of J69591, phenotypically confirmed, in preparation to make glycerol stocks, steak out on a plate, and miniprep. This will be the parent colony for all of our characterizations.
Melanin Retention
Streaks of the strains requested from Philip Oliver containing multi-drug transporter knockouts arrived - MC1060 containg a deletion of the acrAB operon, and MG1655 containing a deletion of the acrF gene (part of the acrEF operon). Competent cell stocks were prepared in readiness for the next day's transformations with the MelA-containing plasmid.
Friday
Wet Work
Melanin Pigment
Made controls (as suggested in the meeting yesterday) to confirm that the melanin pigment was being produced by the bacteria with the MelA gene, rather than by oxidation of plate substrates. Schematic is shown below - all plates contain tyrosine, copper sulfate, and IPTG. Brown colour represents melanin bacteria, while pale stain represents the TOP10 control species. All plates were stored over the weekend at 37 degrees.
Melanin Retention
Electroporation was used to transform the MC1060 (containing an acrAB deletion) and MG1655 (containing an acrF deletion) strains with the MelA-containing plasmid before plating them out onto ampicillin plates. Hopefully these should show limited melanin diffusion into the agar medium and delineated black colonies after a spell in the incubator.
DNA Wash
As part of troubleshooting the column wash procedure, the QUIGEN kit was used to purify the DNA after a PCR. The gel below shoes, from left to right, 2-log ladder, unwashed PCR-product, 1/10 diluted PCR product, and washed DNA. The QUIGEN kit therefore produces a reasonable yield of pure DNA, and will be used in the melanin protocol which will be startedon tuesday.
(GEL PICTURE)
Dry Work
Modelling
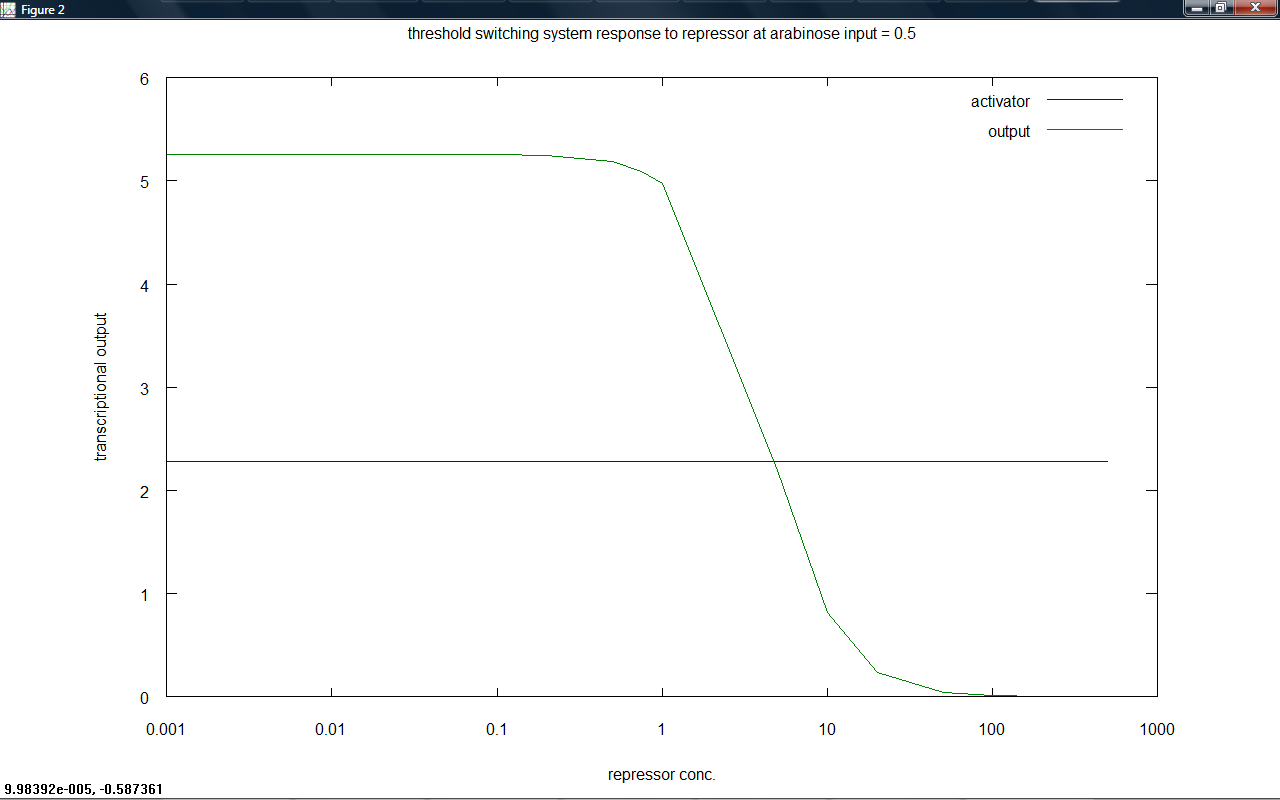
The threshold switching system was modelled, giving model 6.1 (transcription only when activator bound) and model 6.2 (competition between activator and repressor). A plot of output against repressor present is given for a set input which shows the expected switching off of output at a threshold (this is the reverse of an activator switch on, effectively).
Violacein Synthesis
Sent the order to DNA2.0! The GeneDesigner file was send, along with the ApE files for the vio operon and the vector we hope to splice the operon into (pPB3K3). The final design is shown below:
Design specifications were also finalised:
- The gene consists of five open reading frames (vioA-E) each preceded by a ribosome binding site. The whole sequence is held within the BioBrick registry prefix and suffix
- The DNA sequence of the prefix and suffix cannot be altered in any way
- As overexpression of the violacein operon can be toxic for the bacteria, we would like the operon under control of an inducible/repressible promoter, such as the T5 promoter suggested by Jeremy.
- With the exception of the prefix and suffix (which cannot be altered) the following restriction sites are forbidden within the operon: EcoRI, XbaI, SpeI, PstI, NotI
- If possible, we would like the genes to be optimised for E. coli and preferably B. subtilis as well
- In order to easy gene separation, we would like restriction sites on either side of the vio C and the vio D genes. Ideally ones with compatible overhanging ends such as BamHI, BclI and BglII
Vector: pJexpress 401-406 (T5) Cloning Cassette
 "
"