Team:DTU Denmark/USERprinciple
From 2009.igem.org
| Line 1: | Line 1: | ||
| + | [[image:wiki banner_967px.png]] | ||
| + | |||
<html> | <html> | ||
| + | <head> | ||
| + | <title>The project</title> | ||
| + | <style> | ||
| + | #globalWrapper { | ||
| + | background-image: "" !important; | ||
| + | background-repeat: repeat; | ||
| + | |||
| + | } | ||
| + | |||
| + | .ideasList{ | ||
| + | margin-bottom:30px | ||
| + | } | ||
| + | |||
| + | ul#sub { | ||
| + | margin:0px; | ||
| + | padding:0px; | ||
| + | font-size:11px; | ||
| + | list-style-image:none; | ||
| + | list-style-type:none; | ||
| + | |||
| + | } | ||
| + | |||
| + | #sub li { margin-bottom:20px;} | ||
| + | |||
| + | #sub li a { | ||
| + | color:#3300FF; | ||
| + | text-decoration:underline; | ||
| + | } | ||
| + | |||
| + | #sub li a:hover { | ||
| + | text-decoration:none; | ||
| + | } | ||
| + | |||
| + | .links { | ||
| + | width:967px; | ||
| + | background-color:#FFFFFF; | ||
| + | margin-bottom:0px; | ||
| + | } | ||
| + | |||
| + | ..links { | ||
| + | width:967px; | ||
| + | background-color:white; | ||
| + | margin-bottom:0px; | ||
| + | } | ||
| + | |||
| + | .links tr { | ||
| + | background-color:#660066; | ||
| + | height:30px; | ||
| + | } | ||
| + | |||
| + | .links td { | ||
| + | padding: 0; | ||
| + | } | ||
| + | |||
| + | |||
| + | .links a{ | ||
| + | display: block; | ||
| + | width: 130px; | ||
| + | height: 100%; | ||
| + | margin: 0; | ||
| + | text-align: center; | ||
| + | text-decoration: none; | ||
| + | font-size: 15px; | ||
| + | color: #fff; | ||
| + | } | ||
| + | |||
| + | .links a:active { | ||
| + | } | ||
| + | |||
| + | .links a:hover { | ||
| + | background-color: #660066; | ||
| + | } | ||
| + | |||
| + | |||
| + | |||
| + | #header { | ||
| + | width: 100%; | ||
| + | text-align: left; | ||
| + | } | ||
| + | |||
| + | h4 { | ||
| + | font-size: 13pt; | ||
| + | font-weight: normal; | ||
| + | } | ||
| + | |||
| + | dt { | ||
| + | font-size: 12pt; | ||
| + | } | ||
| + | |||
| + | #projectnav h4 { | ||
| + | padding: .4ex; | ||
| + | } | ||
| + | |||
| + | #projectnav dt { | ||
| + | font-weight: normal; | ||
| + | margin: 1.5ex 0 0 0; | ||
| + | } | ||
| + | |||
| + | #projectnav h4, #projectnav dt, #projectnav dd { | ||
| + | text-align: left; | ||
| + | text-indent: 0; | ||
| + | margin-left: 0; | ||
| + | } | ||
| + | |||
| + | #hbnav dd { | ||
| + | padding: .8ex; | ||
| + | } | ||
| + | |||
| + | #denav dd{ | ||
| + | padding: 1ex; | ||
| + | margin-top: 0; | ||
| + | } | ||
| + | |||
| + | #projectnav a { | ||
| + | color: #696969; | ||
| + | } | ||
| + | |||
| + | #pagecontent p { | ||
| + | text-indent: 1.25em; | ||
| + | } | ||
| + | |||
| + | p.start:first-line { | ||
| + | font-weight: bold; | ||
| + | letter-spacing: 1.2; | ||
| + | } | ||
| + | |||
| + | .greybox { | ||
| + | background-color: #DDDDDD; | ||
| + | } | ||
| + | |||
| + | </style> | ||
| + | <! -- END of style sheet --> | ||
| + | |||
| + | </head> | ||
| + | <body> | ||
| + | |||
| + | <!-- First table displays centered iGEM 2009 Logo --> | ||
| + | <div id="header"> | ||
| + | <img src="wiki banner_967px.png" /> | ||
| + | </div> | ||
| + | |||
| + | <!-- Table displaying top row of links --> | ||
| + | <table class="links" > | ||
| + | <tr> | ||
| + | <td align="center" ><font face="arial" size="3"><a class="mainLinks" href="https://2009.igem.org/Team:DTU_Denmark" >Home</a></font> </td> | ||
| + | <td align="center" ><font face="arial" size="3"><a class="mainLinks" href="https://2009.igem.org/Team:DTU_Denmark/team" >The Team</a> </font></td> | ||
| + | <td align="center" ><font face="arial" size="3"><a class="mainLinks" href="https://2009.igem.org/Team:DTU_Denmark/project" >The Project</a> </font></td> | ||
| + | <td align="center" ><font face="arial" size="3"><a class="mainLinks" href="https://2009.igem.org/Team:DTU_Denmark/parts" >Parts submitted</a> </font></td> | ||
| + | <td align="center" ><font face="arial" size="3"><a class="mainLinks" href="https://2009.igem.org/Team:DTU_Denmark/modelling">Modelling</a></font> </td> | ||
| + | <td align="center" ><font face="arial" size="3"><a class="mainLinks" href="https://2009.igem.org/Team:DTU_Denmark/notebook" title="Day to day lab activity">Notebook</a> | ||
| + | |||
| + | </font></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | |||
| + | <table> <!-- Table for content area --> | ||
| + | |||
| + | |||
| + | <!-- Main content area --> | ||
| + | |||
| + | <table cellpadding=10px> | ||
| + | <tr> | ||
| + | <td width="163px" height="100%" valign="top"> | ||
| + | <font color="#990000" face="arial" size="3"> | ||
| + | <br> | ||
| + | The redoxilator<br><br> | ||
| + | </font> | ||
| + | <font color="" face="arial" size="2"> | ||
| + | <a href="https://2009.igem.org/Team:DTU_Denmark/genetic_design" CLASS=leftbar>- Genetic design</a><br> | ||
| + | <a href="https://2009.igem.org/Team:DTU_Denmark/applications" CLASS=leftbar>- Applications and perspectives</a><br> | ||
| + | <a href="https://2009.igem.org/Team:DTU_Denmark/results" CLASS=leftbar>- Results</a><br> | ||
| + | <a href="https://2009.igem.org/Team:DTU_Denmark/safety" CLASS=leftbar>- Safety considerations</a><br><br> | ||
| + | </font> | ||
| + | <font color="#990000" face="arial" size="3"> | ||
| + | <br>The USER assembly standard<br><br> | ||
| + | </font> | ||
| + | <font face="arial" size="2"> | ||
| + | <a href="https://2009.igem.org/Team:DTU_Denmark/USERprinciple" CLASS=leftbar>- USER fusion of biobricks</a><br><br> | ||
| + | </font> | ||
| + | <font color="#990000" face="arial" size="3"> | ||
| + | <br>USER fusion primer design software<br><br> | ||
| + | </font> | ||
| + | <font face="arial" size="2"> | ||
| + | <a href="https://2009.igem.org/Team:DTU_Denmark/USERprogram" CLASS=leftbar>- Abstract</a><br> | ||
| + | <a href="https://2009.igem.org/Team:DTU_Denmark/USERprograminstructions" CLASS=leftbar>- Instructions</a><br> | ||
| + | <a href="https://2009.igem.org/Team:DTU_Denmark/USERprogramoutputformat" CLASS=leftbar>- Output format</a><br> | ||
| + | </font> | ||
| + | </td> | ||
| + | <td width="556px" height="100%" valign="top"> | ||
| + | <font color="#990000" face="arial" size="5"> | ||
| + | <br> | ||
| + | <b>The project</b><br><br><br> | ||
| + | </font> | ||
| + | <font color="#333333" face="arial" size="2.5"> | ||
| + | |||
| + | |||
| + | <!-- INSERT MAIN TEXT HERE! (formatting: <b>bold</> <i>italic> <h4>header</h4>) --> | ||
<p> | <p> | ||
| - | The USER fusion assembly standard allows rapid construction of multi-part devices, without some of the drawbacks of the restriction-enzyme based standard biobrick assembly method. The full USER | + | The USER fusion assembly standard allows rapid construction of multi-part devices, without some of the drawbacks of the restriction-enzyme based standard biobrick assembly method. The full USER assembly standard can be found here: (<a href="http://openwetware.org/wiki/The_BioBricks_Foundation:RFC#BBF_RFC_39:_The_USER_cloning_standard" target="_blank">BBF RFC 39</a>). The main advantages of this assembly method is:<br> |
<br> | <br> | ||
| Line 38: | Line 237: | ||
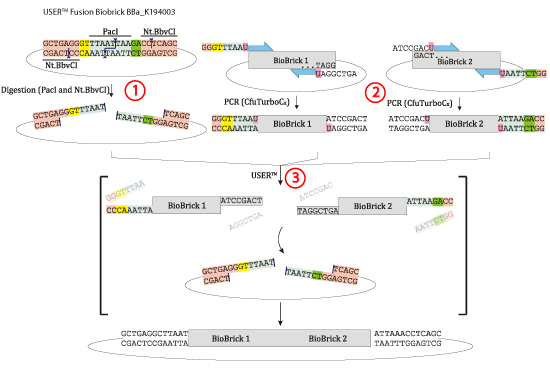
1) The USER fusion biobrick plasmid is digested with the restriction enzyme pacI and the nicking enzyme Nt.BbvCI (a nicking enzyme cuts only one strand as illustrated on the figure). This process will linearize the plasmid, and make single stranded overhangs (sticky ends).<br> | 1) The USER fusion biobrick plasmid is digested with the restriction enzyme pacI and the nicking enzyme Nt.BbvCI (a nicking enzyme cuts only one strand as illustrated on the figure). This process will linearize the plasmid, and make single stranded overhangs (sticky ends).<br> | ||
2) PCR amplification is performed on the biobricks intended for the fusion. The primer design is facilitated by our novel USER fusion primer design software made for this iGEM project.<br> | 2) PCR amplification is performed on the biobricks intended for the fusion. The primer design is facilitated by our novel USER fusion primer design software made for this iGEM project.<br> | ||
| - | 3) USER | + | 3) USER enzyme mix is added. This will remove the uracil always included in the primers, making sticky end overhangs on all biobricks. Because of the matching sticky ends on all biobricks and linearized plamid, the biobricks will self-assemble in the plasmid.<br> |
Two or more biobricks have been joined with all the advantages mentioned above.<br><br> | Two or more biobricks have been joined with all the advantages mentioned above.<br><br> | ||
| - | <a href="http://igem.grafiki.org/" CLASS=leftbar target="blank"> | + | <b>Design your primers with PHUSER <a href="http://igem.grafiki.org/" CLASS=leftbar target="blank">here</a></b><br><br> |
| + | |||
| + | |||
| + | </font> | ||
| + | </td> | ||
| + | |||
| + | <td width="182px" height="100%" valign="top" bgcolor=DDDDDD class="greybox"> | ||
| + | <font face="arial" size="2"> | ||
| + | <b>Synthetic Biology</b><br><br> | ||
| + | |||
| + | |||
| + | <!-- INSERT GREY BOX TEXT HERE! (formatting: <b>bold</> <i>italic> <h4>header</h4>) --> | ||
| + | |||
| + | |||
| + | <p align="left"><i>“Synthetic Biology is an art of engineering new biological systems that don’t exist in nature.”</i><br></p> | ||
| + | |||
| + | <p align="right"><i>-Paras Chopra & Akhil Kamma</i><br><br></p> | ||
| + | |||
| + | <p>In nature, biological molecules work together in complex systems to serve purposes of the cell. In synthetic biology these molecules are used as individual functional units that are combined to form tailored systems exhibiting complex dynamical behaviour. From ‘design specifications’ generated from computational modelling, engineering-based approaches enables the construction of such new specified gene-regulatory networks. The ultimate goal of synthetic biology is to construct systems that gain new functions, and the perspectives of the technology are enormous. It has already been used in several medical projects2 and is predicted to play a major role in biotech-production and environmental aspects.</p> | ||
| + | |||
| + | </font> | ||
| + | </td> | ||
| + | </table> | ||
| + | |||
| + | <table> | ||
| + | |||
| + | <tr height="25px"> | ||
| + | <td width="967px" valign="center" align="center"> | ||
| + | |||
| + | <font face="arial" size="1"> | ||
| + | Comments or questions to the team? Please <a href="mailto:igem@bio.dtu.dk" CLASS=email>Email us</a> -- Comments of questions to webmaster? Please <a href="mailto:lronn@bio.dtu.dk" CLASS=email>Email us</a> | ||
| + | </font> | ||
| + | |||
| + | |||
| + | </td> | ||
| + | </tr> | ||
| + | |||
| + | </table> | ||
| + | |||
| + | </body> | ||
</html> | </html> | ||
Revision as of 21:27, 20 October 2009

| Home | The Team | The Project | Parts submitted | Modelling | Notebook |
|
The redoxilator - Genetic design - Applications and perspectives - Results - Safety considerations The USER assembly standard - USER fusion of biobricks USER fusion primer design software - Abstract - Instructions - Output format |
The project
The USER fusion assembly standard allows rapid construction of multi-part devices, without some of the drawbacks of the restriction-enzyme based standard biobrick assembly method. The full USER assembly standard can be found here: (BBF RFC 39). The main advantages of this assembly method is:
USER fusion of biobricks - how it works
|
Synthetic Biology “Synthetic Biology is an art of engineering new biological systems that don’t exist in nature.” -Paras Chopra & Akhil Kamma In nature, biological molecules work together in complex systems to serve purposes of the cell. In synthetic biology these molecules are used as individual functional units that are combined to form tailored systems exhibiting complex dynamical behaviour. From ‘design specifications’ generated from computational modelling, engineering-based approaches enables the construction of such new specified gene-regulatory networks. The ultimate goal of synthetic biology is to construct systems that gain new functions, and the perspectives of the technology are enormous. It has already been used in several medical projects2 and is predicted to play a major role in biotech-production and environmental aspects. |
| Comments or questions to the team? Please Email us -- Comments of questions to webmaster? Please Email us |
 "
"