Team:Groningen/Modelling/Arsenic
From 2009.igem.org
[http://2009.igem.org/Team:Groningen http://2009.igem.org/wiki/images/f/f1/Igemhomelogo.png]
|
|---|
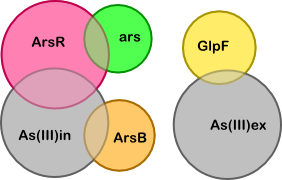
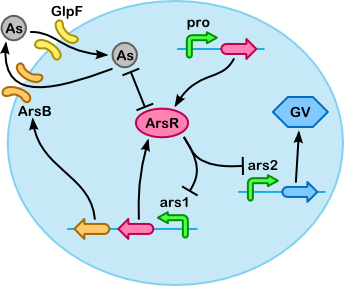
Based on the quasi-steady-state derivation below we have made the simplified version of our model shown below. The simplification is based on two key assumptions (which are also illustrated below, next to the table "Breakdown of core substances"):
- Binding and unbinding of arsenic to/from the transporters occurs on a much smaller time scale than changes in the concentration of arsenic inside and outside the cell. And similarly, we assume that (un)binding of ArsR to/from the ars promoter is much faster than the production of ArsR (for example).
- The concentration of transporters is insignificant compared to the concentration of arsenic inside and outside the cell, and similarly for the concentration of ars promoters compared to the concentration of ArsR. The concentration of promoters might not be insignificant.
This leads to the Michaelis-Menten equation for import, but also some more general equations for export using ArsB and accumulation with ArsR (for example, the Hill equation can be recognized in the activity of the ars promoter).
| Reaction | Description | Rate | |
|---|---|---|---|
| Transport | |||
| As(III)exT → As(III)inT | Import of arsenic. | (Vc/Vs) v5† As(III)exT / (K5+As(III)exT) | |
| As(III)inT → As(III)exT | Export of arsenic. | k8 ArsBAs | |
| ars1T → ars1T + ArsBT | Production of ArsB. | β4 ars1 | |
| ArsBT → null | Degradation of ArsB | ln(2)/τB ArsB | |
| Accumulation | |||
| ars1T → ars1T + ArsRT | Transcription + translation from the chromosomal operon. | β1 ars1 | |
| pro → pro + ArsRT | Transcription + translation from a constitutive promoter. | β3 pro | |
| ArsRT → null | Degradation of ArsR. | (ln(2)/τR) ArsR | |
| Gas vesicles | |||
| ars2T → ars2T + GV | Transcription + translation. | β5 ars2 | |
| GV → null | Degradation of gas vesicles. | ln(2)/τG GV | |
| Name | Description | Derivative to time | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Extracellular | |||||||||||||
| As(III)exT | As(III) in the solution. | (Vc/Vs) k8 ArsBAs - (Vc/Vs) v5† As(III)exT / (K5+As(III)exT) | |||||||||||
| Membrane (all naturally occurring, but we plan to bring GlpF to overexpression) | |||||||||||||
| GlpFT | Importer of As(III) (concentration w.r.t. the exterior of the cell). | (not used directly in model, assumed to be constant) | |||||||||||
| ArsBT | Exporter of As(III) (concentration w.r.t. the interior of the cell). | β4 ars1 - ln(2)/τB ArsB | |||||||||||
| Intracellular (ars2, pro and GV are introduced) | |||||||||||||
| As(III)inT | As(III) (bound and unbound) in the cell. | v5 As(III)exT / (K5+As(III)exT) - k8 ArsBAs | |||||||||||
| ars1T | ArsR repressed promoters (bound and unbound) naturally occurring in E. coli. | (concentration is constant = 1.6605nM, one per cell) | |||||||||||
| ars2T | ArsR repressed promoters in front of gas vesicle genes. | (concentration is constant = 16.605nM, ten per cell) | |||||||||||
| pro | Constitutive promoters in front of arsR. | (concentration is constant = 16.605nM, ten per cell) | |||||||||||
| ArsRT | ArsR in the cell. | β1 ars1 + β3 pro - (ln(2)/τR) ArsR | |||||||||||
| GV | Concentration of gas vesicles. | β5 ars2 - ln(2)/τG GV | |||||||||||
| |||||||||||||
| Name | Units | Value | Description | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| k8 | 1/s | Reaction rate constant representing how fast ArsB can export arsenic. | |||||||||||
| K1d | M | 6µM | Dissociation constant for ArsR and As(III). Assumed to be about an order of magnitude smaller than K2d = 60µM, the corresponding constant for the similar protein ArsD from Chen1997. | ||||||||||
| K3d | M | (0.33µM)² | Dissociation constants for ArsR and ars.
| ||||||||||
| v5 | mol/(s·L) | 3.1863µmol/(s·L) | Maximum import rate per liter of cells (see Michaelis-Menten equation). Note that we have purposefully chosen to write the units as mol/(s·L) instead of M/s, to emphasize the fact that the rate is per liter of cells.
| ||||||||||
| K5 | M | 27.718µM | Concentration at which import reaches half its maximum import rate (see Michaelis-Menten equation).
| ||||||||||
| K7 | M | Concentration at which export reaches half its maximum export rate (see Michaelis-Menten equation).
| |||||||||||
| τB, τR, τG | s | Half-lifes (of ArsB, ArsR and GV, respectively). Degradation rate = ln(2)/τ If you take just the degradation into account you will have the equation dC/dt = -k*C, which leads to C(t) = C(0) e-k t. So if k = ln(2)/τ we get C(t) = C(0) e-ln(2)/τ t = C(0) 2-t/τ. In other words τ is the time it takes for the concentration to half. i | |||||||||||
| β1, β2, etc. | 1/s | Production rates.
| |||||||||||
| Vs | L | Volume of solution (excluding cells). | |||||||||||
| Vc | L | Total volume of cells (in solution) (so Vs+Vc is the total volume). | |||||||||||
| |||||||||||||
The raw model
The following variables play an important role in our system (these can be concentrations of substances, the density of the cell, etc.):
- Extracellular:
- As(III)ex
- Membrane (all naturally occurring, but we plan to bring GlpF to overexpression):
- GlpF (concentration w.r.t. the exterior of the cell)
- GlpFAs (concentration w.r.t. the exterior of the cell)
- ArsB (concentration w.r.t. the interior of the cell)
- ArsBAs (concentration w.r.t. the interior of the cell)
- Intracellular (ars2, pro and GV are introduced):
- As(III)ex
- ars1 (concentration of unbound promoters naturally occurring in E. coli)
- ars2 (concentration of unbound promoters in front of gas vesicle genes)
- proR (concentration of constitutive promoters in front of arsR)
- proM (concentration of constitutive promoters in front of mbp-arsR)
- proF (concentration of constitutive promoters in front of fMT)
- ArsR ArsR binds to ars to repress production of the genes they regulate, and binds to As(III) to make it less of a problem for the cell.i
- ArsRAs (bound to As(III))
- ArsRars1 (bound to ars1)
- ArsRars2 (bound to ars2)
- MBPArsR A fusion of maltose binding protein and ArsR.i
- MBPArsRAs (bound to As(III))
- fMT
- fMTAs (bound to multiple As(III))
- ArsRAs (bound to As(III))
- GV (concentration of gas vesicles)
The variables above can be related to each other through the following "reactions" (color coding is continued below to show which parts of the differential equations refer to which groups of reactions):
- Transport (based on Rosen1996, Meng2004 and Rosen2009)
- As(III)ex + GlpF ↔ GlpFAs
- GlpFAs → GlpF + As(III)
- As(III)in + ArsB ↔ ArsBAs
- ArsBAs → ArsB + As(III)ex
- ArsB → null (degradation)
- Accumulation (mostly based on Chen1997)
- As(III)in + ArsR ↔ ArsRAs
- As(III)in + MBPArsR ↔ MBPArsRAs
- nf As(III)in + fMT ↔ fMTAs
- ars1 + 2 ArsR ↔ ArsRars1
- ars2 + 2 ArsR ↔ ArsRars2
- ars1 → ars1 + ArsR + ArsB (transcription + translation)
- ars2 → ars2 + GV (transcription + translation)
- proR → proR + ArsR (transcription + translation)
- proM → proM + MBPArsR (transcription + translation)
- proF → proF + fMT (transcription + translation)
- ArsR → null (degradation)
- GV → null (degradation)
Resulting in the following differential equations (please note that some can be formed by linear combinations of the others), using color coding to show the correspondence to the reactions above:
- (d/dt) As(III)ex = - (d/dt) GlpFAs - k6 GlpFAs + (Vc/Vs) k8 ArsBAs
- (d/dt) GlpF = - (d/dt) GlpFAs
- (d/dt) GlpFAs = k5on As(III)ex GlpF - (k5off+k6) GlpFAs
- (d/dt) ArsB = - (d/dt) ArsBAs + β4 ars1 - ln(2)/τB ArsB
- (d/dt) ArsBAs = k7on As(III)in ArsB - (k7off+k8) ArsBAs
- (d/dt) As(III)in = - (d/dt) ArsRAs - (d/dt) MBPArsRAs - nf (d/dt) fMTAs - (d/dt) ArsBAs - k8 ArsBAs + (Vs/Vc) k6 GlpFAs
- (d/dt) ars1 = - (d/dt) ArsRars1
- (d/dt) ars2 = - (d/dt) ArsRars2
- (d/dt) ArsR = βRN ars1 + βR proR - (ln(2)/τR) ArsR - (d/dt) ArsRAs - 2 (d/dt) ArsRars1 - 2 (d/dt) ArsRars2
- (d/dt) ArsRAs = kRon ArsR As(III)in - kRoff ArsRAs
- (d/dt) ArsRars1 = k3on ArsR² ars1 - k3off ArsRars1
- (d/dt) ArsRars2 = k3on ArsR² ars2 - k3off ArsRars2
- (d/dt) MBPArsR = βM proM - (ln(2)/τM) MBPArsR - (d/dt) MBPArsRAs
- (d/dt) MBPArsRAs = kMon MBPArsR As(III)in - kMoff MBPArsRAs
- (d/dt) fMT = βF proF - (ln(2)/τF) fMT - (d/dt) fMTAs
- (d/dt) fMTAs = kFon fMT As(III)innf - kFoff fMTAs
- (d/dt) GV = βG ars2 - ln(2)/τG GV
Using the following constants/definitions:
| Name | Units | Description |
|---|---|---|
| kRon, kMon, k5on, etc. | 1/(M·s) | Reaction rate constants for reactions to a complex. |
| k3on | 1/(M²·s) | Reaction rate constants for reactions to a complex. |
| kFon | 1/(Mnf·s) | Reaction rate constants for reactions to a complex. |
| kRoff, kMoff, kFoff, k3off, k5off, etc. | 1/s | Reaction rate constants for reactions from a complex. |
| k6, k8 | 1/s | Reaction rate constants representing how fast transporters transport their cargo to "the other side". |
| τB, τR, τM, τF, τG | s | Half-lifes (of ArsB, ArsR, MBPArsR, fMT and GV, respectively). Degradation rate = ln(2)/τ If you take just the degradation into account you will have the equation dC/dt = -k*C, which leads to C(t) = C(0) e-k t. So if k = ln(2)/τ we get C(t) = C(0) e-ln(2)/τ t = C(0) 2-t/τ. In other words τ is the time it takes for the concentration to half. i |
| βRN, βR, etc. | 1/s | Production rates.
|
| Vs | L | Volume of solution (excluding cells). |
| Vc | L | Total volume of cells (in solution) (so Vs+Vc is the total volume). |
See Chen1997 for the interplay between ArsR and ArsD (the latter has a role similar to ArsR, but we do not treat it, as it is not present in our system).
Quasi steady state
First of all, we assume the concentration of transporters is quite low compared to the concentration of the transported substances. After all, if this were not the case the transporters would act more like "storage" proteins than transporters (note that this can be even more rigorously justified if, for example, GlpFT<<K5). Similarly, there will generally only be a few ars promoters, compared to very many ArsR molecules. This leads to:
As(III)exT ≈ As(III)ex As(III)inT ≈ As(III)in + ArsRAs + MBPArsRAs + fMTAs ArsRT ≈ ArsR + ArsRAs
Also, we assume the binding and unbinding of molecules to the transporters occurs on a much finer time-scale than any actual changes to the concentrations inside and outside the cell. Similarly, within the cell we assume diffusion processes are very fast and binding/unbinding of substances is quite fast compared to the production of proteins. This leads us to assume that the following ratios between substances are constantly in equilibrium:
As(III)ex : GlpFAs ≈ As(III)ex : 0 GlpF : GlpFAs ArsB : ArsBAs As(III)in : ArsRAs : MBPArsRAs : fMTAs : ArsBAs ≈ As(III)in : ArsRAs : MBPArsRAs : fMTAs : 0 ArsR : ArsRAs : 2 ArsRars ≈ ArsR : ArsRAs : 0 ars : ArsRars
To determine what the unknown ratios are we can set the following derivatives to zero (these are the derivatives of the complexes corresponding to the four overlapping regions in the diagram):
0 = (d/dt) GlpFAs = k5on As(III)ex GlpF - (k5off+k6) GlpFAs 0 = (d/dt) ArsBAs = k7on As(III)in ArsB - (k7off+k8) ArsBAs 0 = (d/dt) ArsRars = k3on ArsR² ars - k3off ArsRars 0 = (d/dt) ArsRAs = kRon ArsR As(III)in - kRoff ArsRAs 0 = (d/dt) MBPArsRAs = kMon MBPArsR As(III)in - kMoff MBPArsRAs 0 = (d/dt) fMTAs = kFon fMT As(III)in^nf - kFoff fMTAs
The first two derivates let us determine the ratios between bound and unbound transporters:
0 = (d/dt) GlpFAs = k5on As(III)ex GlpF - (k5off+k6) GlpFAs
k5on As(III)ex GlpF = (k5off+k6) GlpFAs
GlpF = (k5off+k6)/k5on GlpFAs / As(III)ex
GlpF = K5 GlpFAs / As(III)ex
GlpF : GlpFAs
K5 GlpFAs / As(III)ex : GlpFAs
K5 : As(III)ex
0 = (d/dt) ArsBAs = k7on As(III)in ArsB - (k7off+k8) ArsBAs
k7on As(III)in ArsB = (k7off+k8) ArsBAs
ArsB = (k7off+k8)/k7on ArsBAs / As(III)in
ArsB = K7 ArsBAs / As(III)in
ArsB : ArsBAs
K7 ArsBAs / As(III)in : ArsBAs
K7 : As(III)in
The next two differential equations can be used to determine the relative abundances of ArsR and ArsRAs, and ars and ArsRars:
0 = (d/dt) ArsRAs = kRon ArsR As(III)in - kRoff ArsRAs
kRon ArsR As(III)in = kRoff ArsRAs
ArsRAs = kRon/kRoff ArsR As(III)in
ArsRAs = ArsR As(III)in / KRd
ArsR : ArsRAs
ArsR : ArsR As(III)in / KRd
KRd : As(III)in
0 = (d/dt) ArsRars = k3on ArsR² ars - k3off ArsRars
k3on ArsR² ars = k3off ArsRars
ArsRars = k3on/k3off ArsR² ars
ArsRars = ArsR² ars / K3d²
ars : ArsRars
ars : ArsR² ars / K3d²
K3d² : ArsR²
For MBPArsR and fMT we find:
0 = (d/dt) MBPArsRAs = kMon MBPArsR As(III)in - kMoff MBPArsRAs MBPArsR : MBPArsRAs = KMd : As(III)in 0 = (d/dt) fMTAs = kFon fMT As(III)in^nf - kFoff fMTAs fMT : fMTAs = KFd^nf : As(III)in^nf
And finally the relative abundances of arsenic:
ArsRAs = ArsR As(III)in / K1d As(III)in : ArsRAs : MBPArsRAs : fMTAs As(III)in : ArsR As(III)in / KRd : MBPArsR As(III)in / KMd : fMT As(III)in^nf / KFd^nf 1 : ArsR / KRd : MBPArsR / KMd : fMT As(III)in^(nf-1) / KFd^nf
Summarizing:
As(III)ex : GlpFAs ≈ As(III)ex : 0 GlpF : GlpFAs = K5 : As(III)ex ArsB : ArsBAs = K7 : As(III)in As(III)in : ArsRAs : MBPArsRAs : fMTAs ≈ 1 : ArsR / KRd : MBPArsR / KMd : fMT As(III)in^(nf-1) / KFd^nf ars : ArsRars = K3d² : ArsR² ArsR : ArsRAs ≈ K1d : As(III)in MBPArsR : MBPArsRAs = KMd : As(III)in fMT : fMTAs = KFd^nf : As(III)in^nf
Now we can look at the differential equations for the totals of ArsB (so ArsBT=ArsB+ArsBAs), ArsR, As(III)in and As(III)ex (GlpFT and arsT are assumed to be constant):
(d/dt) As(III)exT = (d/dt) As(III)ex + (d/dt) GlpFAs
= - (d/dt) GlpFAs - k6 GlpFAs + (Vc/Vs) k8 ArsBAs + (d/dt) GlpFAs
= (Vc/Vs) k8 ArsBAs - k6 GlpFAs
= (Vc/Vs) k8 ArsBAs - (Vc/Vs) v5 GlpFAs / GlpFT
= (Vc/Vs) k8 ArsBAs - (Vc/Vs) v5 As(III)ex / (K5+As(III)ex)
= (Vc/Vs) k8 ArsBAs - (Vc/Vs) v5 As(III)exT / (K5+As(III)exT)
(d/dt) ArsBT = (d/dt) ArsB + (d/dt) ArsBAs
= - (d/dt) ArsBAs + β4 ars1 - ln(2)/τB ArsB + (d/dt) ArsBAs
= β4 ars1 - ln(2)/τB ArsB
(d/dt) As(III)inT = -(Vs/Vc) (d/dt) As(III)exT
= v5 As(III)exT / (K5+As(III)exT) - k8 ArsBT As(III)in / (K7+As(III)in)
(d/dt) ArsRT = (d/dt) ArsR + (d/dt) ArsRAs + 2 (d/dt) ArsRars
= βRN ars1 + βR proR - (ln(2)/τR) ArsR - (d/dt) ArsRAs - 2 (d/dt) ArsRars + (d/dt) ArsRAs + 2 (d/dt) ArsRars
= βRN ars1 + βR proR - (ln(2)/τR) ArsR
(d/dt) MBPArsRT = (d/dt) MBPArsR + (d/dt) MBPArsRAs
= βM proM - (ln(2)/τM) MBPArsR
(d/dt) fMTT = (d/dt) fMT + (d/dt) fMTAs
= βF proF - (ln(2)/τF) fMT
Steady state
By looking at the steady state of the system we can say something about its long-term behaviour. This also makes it easier to analyze relations between variables. To derive the steady state solution we take the quasi steady state solution and simplify it further by setting additional derivatives to zero:
0 = (d/dt) ArsBT = β4 ars1 - ln(2)/τB ArsB 0 = (d/dt) As(III)inT = v5 As(III)exT / (K5+As(III)exT) - k8 ArsBAs 0 = (d/dt) ArsRT = β1 ars1 + β3 pro - (ln(2)/τR) ArsR 0 = (d/dt) GV = β5 ars2 - ln(2)/τG GV
This directly leads to:
0 = β4 ars1 - ln(2)/τB ArsB ArsB = β4 (τB/ln(2)) ars1 ArsB = β4 (τB/ln(2)) ars1T K3d²/(K3d²+ArsR²) 0 = β5 ars2 - ln(2)/τG GV GV = β5 (τB/ln(2)) ars2 GV = β5 (τB/ln(2)) ars2T K3d²/(K3d²+ArsR²)
For the intra- and extracellular concentrations we can find the following condition:
0 = v5 As(III)exT / (K5+As(III)exT) - k8 ArsBAs 0 = v5 As(III)exT / (K5+As(III)exT) - k8 ArsB As(III)in / K7 0 = v5 As(III)exT / (K5+As(III)exT) - k8 ArsB As(III)inT / (K7 (1+ArsR/K1d)) 0 = v5 As(III)exT (K7(1+ArsR/K1d)) - k8 ArsB As(III)inT (K5+As(III)exT) 0 = v5 K7 (1+ArsR/K1d) As(III)exT - k8 ArsB K5 As(III)inT - k8 ArsB As(III)exT As(III)inTAs we can safely assume arsenic neither disappears into nothingness nor appears from nothingness, we can use this to derive a quadratic equation for As(III)inT
b² ≥ 4 a c. This would allow both a plus and a minus to yield positive solutions, however, by also examining the upper bound Vc As(III)inT ≤ As(III)T it can be shown that only the minus sign is appropriate. In addition, the alternative form of the solution is algebraically equivalent and prevents numerical problems for a→0.
As(III)T = Vs As(III)exT + Vc As(III)inT
Vs As(III)exT = As(III)T - Vc As(III)inT
0 = v5 K7 (1+ArsR/K1d) As(III)exT - k8 ArsB K5 As(III)inT - k8 ArsB As(III)exT As(III)inT
0 = v5 K7 (1+ArsR/K1d) Vs As(III)exT - k8 ArsB Vs K5 As(III)inT - k8 ArsB Vs As(III)exT As(III)inT
0 = v5 K7 (1+ArsR/K1d) (As(III)T - Vc As(III)inT) - k8 ArsB Vs K5 As(III)inT - k8 ArsB (As(III)T - Vc As(III)inT) As(III)inT
0 = v5 K7 (1+ArsR/K1d) As(III)T - v5 Vc K7 (1+ArsR/K1d) As(III)inT - k8 ArsB Vs K5 As(III)inT - k8 ArsB As(III)T As(III)inT + k8 ArsB Vc As(III)inT²
0 = v5 K7 (1+ArsR/K1d) As(III)T - (v5 Vc K7 (1+ArsR/K1d) + k8 ArsB Vs K5 + k8 ArsB As(III)T) As(III)inT + k8 ArsB Vc As(III)inT²
0 = v5 K7 (1+ArsR/K1d) As(III)T - (v5 Vc K7 (1+ArsR/K1d) + k8 ArsB (Vs K5 + As(III)T)) As(III)inT + k8 ArsB Vc As(III)inT²
As(III)inT = (b - √(b² - 4 a c))/(2 a)
= 2 c/(b + √(b² - 4 a c))
a = k8 ArsB Vc
b = v5 Vc K7 (1+ArsR/K1d) + k8 ArsB (Vs K5 + As(III)T)
c = v5 K7 (1+ArsR/K1d) As(III)T
Finally, for ArsR we can find the following third-order equation:
0 = β1 ars1 + β3 pro - (ln(2)/τR) ArsR 0 = β1 ars1T K3d²/(K3d²+ArsR²) + β3 pro - (ln(2)/τR) ArsR 0 = β1 ars1T K3d² + β3 pro (K3d²+ArsR²) - (ln(2)/τR) ArsR (K3d²+ArsR²) 0 = β1 ars1T K3d² + β3 pro K3d² + β3 pro ArsR² - (ln(2)/τR) ArsR K3d² - (ln(2)/τR) ArsR³ 0 = (β1 ars1T + β3 pro) K3d² - (ln(2)/τR) K3d² ArsR + β3 pro ArsR² - (ln(2)/τR) ArsR³ 0 = (β1 ars1T + β3 pro) (τR/ln(2)) K3d² - K3d² ArsR + β3 (τR/ln(2)) pro ArsR² - ArsR³
According to Mathematica's solution of Reduce[eq && K3d > 0 && arsT >= 0 && pro >= 0 && β1 > 0 && β3 > 0 && τR > 0, ArsR, Reals] (where eq is the equation shown above) there is only one real solution (examining the discriminant of eq confirms this), so we can solve the equation safely using Newton's (or Halley's) method.
 "
"