Team:Groningen/Modelling/Arsenic
From 2009.igem.org
Bold text
[http://2009.igem.org/Team:Groningen http://2009.igem.org/wiki/images/f/f1/Igemhomelogo.png]
|
|---|
Our initial ideas on how and what to model can be found at Brainstorm/Modelling.
Usage of graphs in wiki: Graphs
The raw model
Note: Math support is currently not enabled on this Wiki... (I've asked hq if they can enable it.)
The following variables play an important role in our system (these can be concentrations of substances, the density of the cell, etc.):
- Extracellular:
- As(III)ex
-
As(V)ex
- Membrane:
- GlpF
- GlpFAs
- Intracellular:
- As(III)
- OpN (concentration of unbound operators, not used in this model)
- OpG (concentration of unbound operators)
- OpH (concentration of ArsR producing operators that are always on)
-
As(V) -
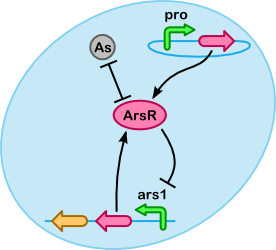
ArsC - ArsR ArsR binds to OpN/OpG to repress production of the genes they regulate, and binds to As(III) to make it less of a problem for the cell.i
- ArsD
- ArsRAs (bound to As(III))
- At equilibrium: ArsR As(III) = (k1off/k1on) ArsRAs
- ArsDAs (bound to As(III))
- ArsRopn (bound to operator)
- ArsDopn (bound to operator)
- ArsRopg (bound to opg)
- ArsDopg (bound to opg)
The variables above can be related to each other through the following "reactions" and/or equations:
- Transport
-
As(V)ex → As(V), using phosphate transporters? (Summers2009) -
As(V)ex → As(III), using ArsC (Summers2009) -
As(III) → As(III)ex, using ArsAB (helped by ArsD) (Summers2009) - As(III)ex + GlpF ↔ GlpFAs
- GlpFAs → GlpF + As(III)
-
- Accumulation
- As(III)+ ArsR ↔ ArsRAs
- As(III) + ArsD ↔ ArsDAs
- OpN + ArsR ↔ ArsRopn
- OpN + ArsD ↔ ArsDopn
- OpG + ArsR ↔ ArsRopg
- OpG + ArsD ↔ ArsDopg
- OpN → OpN + ArsR + ArsD (transcription + translation)
- OpG → OpG + ArsR (transcription + translation)
- OpH → OpH + ArsR (transcription + translation)
- ArsR → null (degradation)
- ArsD → null (degradation)
Resulting in the following differential equations (please note that some can be formed by linear combinations of the others):
- (d/dt) As(III)ex = -k5on As(III)ex GlpF + k5off GlpFAs
- (d/dt) GlpF = -k5on As(III)ex GlpF + (k5off+k6) GlpFAs
- (d/dt) GlpFAs = k5on As(III)ex GlpF - (k5off+k6) GlpFAs
- (d/dt) As(III) = - (k1on ArsR+k2on ArsD) As(III) + k1off ArsRAs + k2off ArsDAs + k6 GlpFAs
- (d/dt) OpN = - (k3on ArsR+k4on ArsD) OpN + k3off ArsRopn + k3off ArsDopn
- (d/dt) OpG = - (k3on ArsR+k4on ArsD) OpG + k3off ArsRopg + k3off ArsDopg
- (d/dt) ArsR = β1 (OpN+OpG) + β3 OpH - (ln(2)/τ1+k1on As(III)+k3on (OpN+OpG)) ArsR + k1off ArsRAs + k3off (ArsRopn+ArsRopg)
- (d/dt) ArsD = β2 OpN - (ln(2)/τ2+k2on As(III)+k4on (OpN+OpG)) ArsD + k2off ArsDAs + k4off (ArsDopn+ArsDopg)
- (d/dt) ArsRAs = k1on ArsR As(III) - k1off ArsRAs
- (d/dt) ArsDAs = k2on ArsD As(III) - k2off ArsDAs
- (d/dt) ArsRopn = k3on ArsR OpN - k3off ArsRopn
- (d/dt) ArsDopn = k4on ArsD OpN - k4off ArsDopn
- (d/dt) ArsRopg = k3on ArsR OpG - k3off ArsRopg
- (d/dt) ArsDopg = k4on ArsD OpG - k4off ArsDopg
Using the following constants/definitions:
- K1d = k1off/k1on
- K2d = k2off/k2on = 60µM (Chen1997)
- K3d = k3off/k3on = 0.33µM (Chen1997, suspect as the relevant reference doesn't actually seem to give any value for this)
- K4d = k4off/k4on = 65µM (Chen1997)
- degradation rate = ln(2)/τ
- ArsR half-life time = τ1
- ArsD half-life time = τ2
- β1 = β2 ??? (and either value is unknown)
- β2, the production rate for ArsR behind our constitutive promotor
See Chen1997 for the interplay between ArsR and ArsD.
TODO Figure out relevant equations for metallochaperone function of ArsD?
TODO Make sure all the multiplicities are correct (and/or taken care of in constants). E.g. does 1 mol ArsR (if it is bound) bind 1 mol As(III)?
Equilibrium
For many purposes, like determining the total amount of accumulated arsenic, it can be quite useful to consider the system at equilibrium. That is, when the derivatives of all variables to time are zero (just taking the equations relevant for accumulation here, assuming the As(III) concentration to be constant):
0 = - (k1on ArsR+k2on ArsD) As(III) + k1off ArsRAs + k2off ArsDAs 0 = - (k3on ArsR+k4on ArsD) OpN + k3off ArsRopn + k3off ArsDopn 0 = - (k3on ArsR+k4on ArsD) OpG + k3off ArsRopg + k3off ArsDopg 0 = β1 (OpN+OpG) + β3 OpH - (ln(2)/τ1+k1on As(III)+k3on (OpN+OpG)) ArsR + k1off ArsRAs + k3off (ArsRopn+ArsRopg) 0 = β2 OpN - (ln(2)/τ2+k2on As(III)+k4on (OpN+OpG)) ArsD + k2off ArsDAs + k4off (ArsDopn+ArsDopg) 0 = k1on ArsR As(III) - k1off ArsRAs 0 = k2on ArsD As(III) - k2off ArsDAs 0 = k3on ArsR OpN - k3off ArsRopn 0 = k4on ArsD OpN - k4off ArsDopn 0 = k3on ArsR OpG - k3off ArsRopg 0 = k4on ArsD OpG - k4off ArsDopg
From the last four equations it can be seen that the ratio between OpN and ArsRopn should be equal to the ratio between OpG and ArsRopg, and similarly for ArsDop?. This leads to:
Op = OpN + OpG
OpN/OpG = OpNT/OpGT
OpN/Op = OpNT/OpT
ArsRop = ArsRopn + ArsRopg
ArsDop = ArsDopn + ArsDopg
0 = - (k1on ArsR+k2on ArsD) As(III) + k1off ArsRAs + k2off ArsDAs
0 = - (k3on ArsR+k4on ArsD) Op + k3off ArsRop + k3off ArsDop
0 = β1 Op + β3 OpH - (ln(2)/τ1+k1on As(III)+k3on Op) ArsR + k1off ArsRAs + k3off ArsRop
0 = β2 OpN - (ln(2)/τ2+k2on As(III)+k4on Op) ArsD + k2off ArsDAs + k4off ArsDop
0 = k1on ArsR As(III) - k1off ArsRAs
0 = k2on ArsD As(III) - k2off ArsDAs
0 = k3on ArsR Op - k3off ArsRop
0 = k4on ArsD Op - k4off ArsDop
By eliminating the last four equations from the rest and dividing the last four by k?on we are left with:
0 = β1 Op + β3 OpH - (ln(2)/τ1) ArsR 0 = β2 OpN - (ln(2)/τ2) ArsD 0 = ArsR As(III) - K1d ArsRAs 0 = ArsD As(III) - K2d ArsDAs 0 = ArsR Op - K3d ArsRop 0 = ArsD Op - K4d ArsDop
Using the fact that the total amount of operators remains constant the last two equations can be used to derive an equation for Op:
ArsRop = Op ArsR / K3d ArsDop = Op ArsD / K4d OpT = Op + ArsRop + ArsDop OpT = Op + Op ArsR / K3d + Op ArsD / K4d OpT = Op (1 + ArsR/K3d + ArsD/K4d) Op = OpT/(1 + ArsR/K3d + ArsD/K4d)
This leads to the following:
0 = β1 Op + β3 OpH - (ln(2)/τ1) ArsR 0 = β1 OpT + (β3 OpH - (ln(2)/τ1) ArsR) (ArsR/K3d + ArsD/K4d + 1) 0 = β1 OpT + β3 OpH ArsR/K3d + β3 OpH (ArsD/K4d + 1) - (ln(2)/τ1) ArsR²/K3d - (ln(2)/τ1) ArsR ArsD/K4d - (ln(2)/τ1) ArsR 0 = K3d (τ1/ln(2)) β1 OpT + (τ1/ln(2)) β3 OpH ArsR + K3d (τ1/ln(2)) β3 OpH (ArsD/K4d + 1) - ArsR² - (K3d/K4d) ArsD ArsR - K3d ArsR 0 = ½ ArsR² + ½ (K3d (ArsD/K4d + 1) - (τ1/ln(2)) β3 OpH) ArsR - ½ K3d (τ1/ln(2)) (β1 OpT + β3 OpH (ArsD/K4d + 1)) ArsR = -b1 ± √(b1² + c1) b1 = ½ (K3d (ArsD/K4d + 1) - (τ1/ln(2)) β3 OpH) c1 = K3d (τ1/ln(2)) (β1 OpT + β3 OpH (ArsD/K4d + 1)) 0 = β2 OpN - (ln(2)/τ2) ArsD 0 = β2 OpNT - (ln(2)/τ2) ArsD (ArsR/K3d + ArsD/K4d + 1) ArsD = -b2 ± √(b2² + c2) b2 = ½ K4d (ArsR/K3d + 1) c2 = K4d (τ2/ln(2)) β2 OpNT
As b2 is positive (concentrations are always non-negative) only the plus sign of the plus-minuses in the equation for ArsD is a valid choice. In addition, since c1 and c2 are non-negative the square roots are always larger than or equal to the magnitude of b1/b2, so the solution will be non-negative if and only if a plus is used in both equations. These equations for ArsR and ArsD can now be used in a fixed point iteration as follows:
f(ArsR) = -b1 + √(b1² + c1) with ArsD = -b2 + √(b2² + c2) f' = -b1' + (2 b1 b1' + c1')/√(b1² + c1) b1' = ½ (K3d/K4d) ArsD' c1' = (τ1/ln(2)) β3 OpH (K3d/K4d) ArsD' ArsD' = -b2' + (2 b2 b2')/√(b2² + c2) b2' = ½ (K4d/K3d)
Analogously to the derivation of the equation for Op an equation can be derived for the fraction of unbound arsenic in the cell:
As(III)T = As(III) + ArsRAs + ArsDAs As(III)/As(III)T = 1/(ArsR/K1d + ArsD/K2d + 1)
Kinetic Laws
TODO Add references.
TODO Find out how to determine experimentally which is applicable (and if you know, what the parameters are).
- Mass Action
- Molecules randomly interact, the reaction rate is simply the product of the concentrations of the reactants (multiplied by a constant).
- Michaelis-Menten
- Applicable to situations where there is a maximum reaction rate (due to needing a catalyst/transporter/binding site of which there is only a limited amount for example) under the assumption that there is much more of the "main" reactant than of the catalyst/transporter. Has two constants, the maximum reaction rate and the concentration at which the reaction rate is half the maximum reaction rate.
- Michaelis-Menten reversible
- TODO
- Hill
- Generalization of Michaelis-Menten. More detail.
For rate parameters it is best to have both the forward and reverse reaction rates, if you don't then a dissociation constant can be used (which is the ratio of the reverse and forward rates), in combination with a "standard" rate of 108-109 (see appendix A of Alon2007), in the case of two reactants at least.
See http://www.biomodels.net/ for a database of models.
Protein production
Concepts:
- translation rate
- Number of amino acids per second that are translated (per mRNA). Expressed in amino acids per second per mRNA?
- ribosome occupancy
- Fraction of mRNA bound to ribosomes.
- mRNA abundance
- Amount of mRNA (molecules per cell or concentration).
- translational activity / protein production rate ?
- Translation rate per mRNA times mRNA abundance (units?: 1/(mol*sec) * mol/L = 1/(sec*L)). ???
 "
"