Team:Groningen/Project/WholeSystem
From 2009.igem.org
[http://2009.igem.org/Team:Groningen http://2009.igem.org/wiki/images/f/f1/Igemhomelogo.png]
|
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Introduction
Cloning Strategy
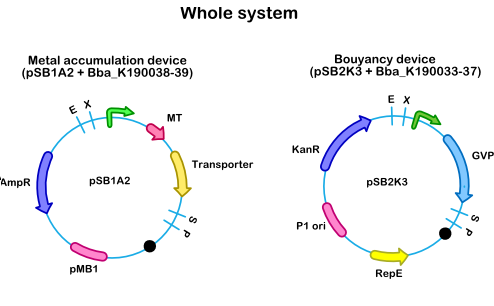
- Figure 1: Whole system, combining the bouyancy device on a pSB2K3 vector and the accumulation device on a pSB1AC3 vector
Results
Combining parts and was done by using a normal transformation protocol with both ampicillin and kanamycin as antibiotics. A buoyancy test was performed as described also using both ampicillin and kanmycin as antibiotics. Iptg was also added to the dayculture to induce the part. In exponential phase 10μM NaAsO2 was added to half of the samples. A restriction was done to check the transformation.
Restriction
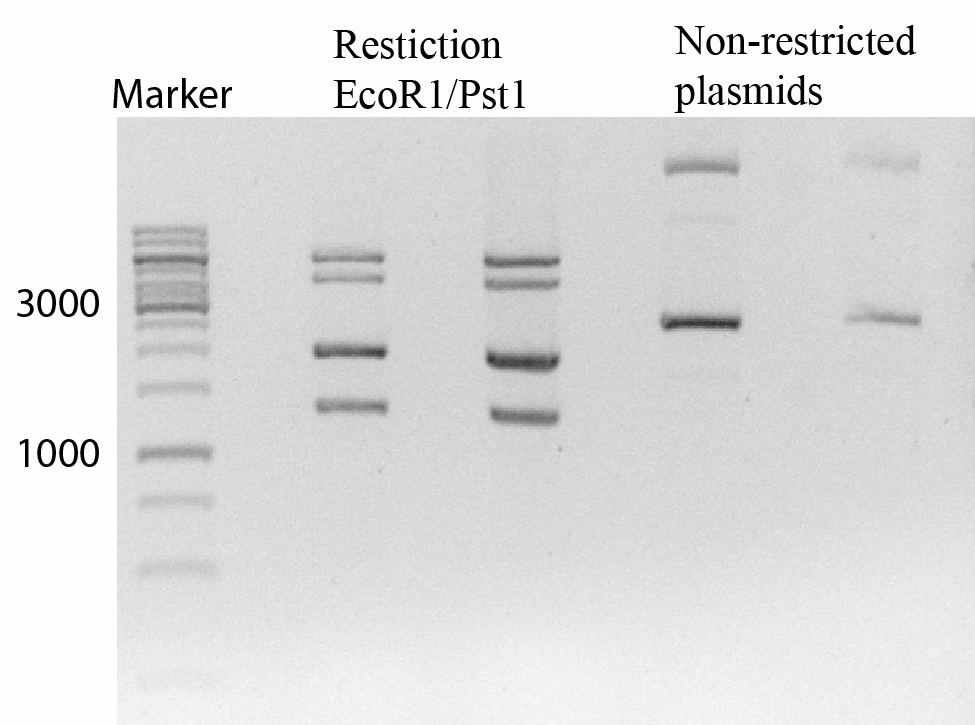
Figure 1 shows that the transformation of both vectors succeeded.
Figure 1
The gel shows a 1Kb marker to the far left, after that two times a restriction with EcoRI and PstI can be seen. This shows clearly fragments of 6000pb and 4500pb for the and fragments of 2000pb and 1300 bp for the . The next two slots show non-restricted plasmids, two distinc bands can be seen clearly indicating the presence of two plasmids.
Buoyancy test
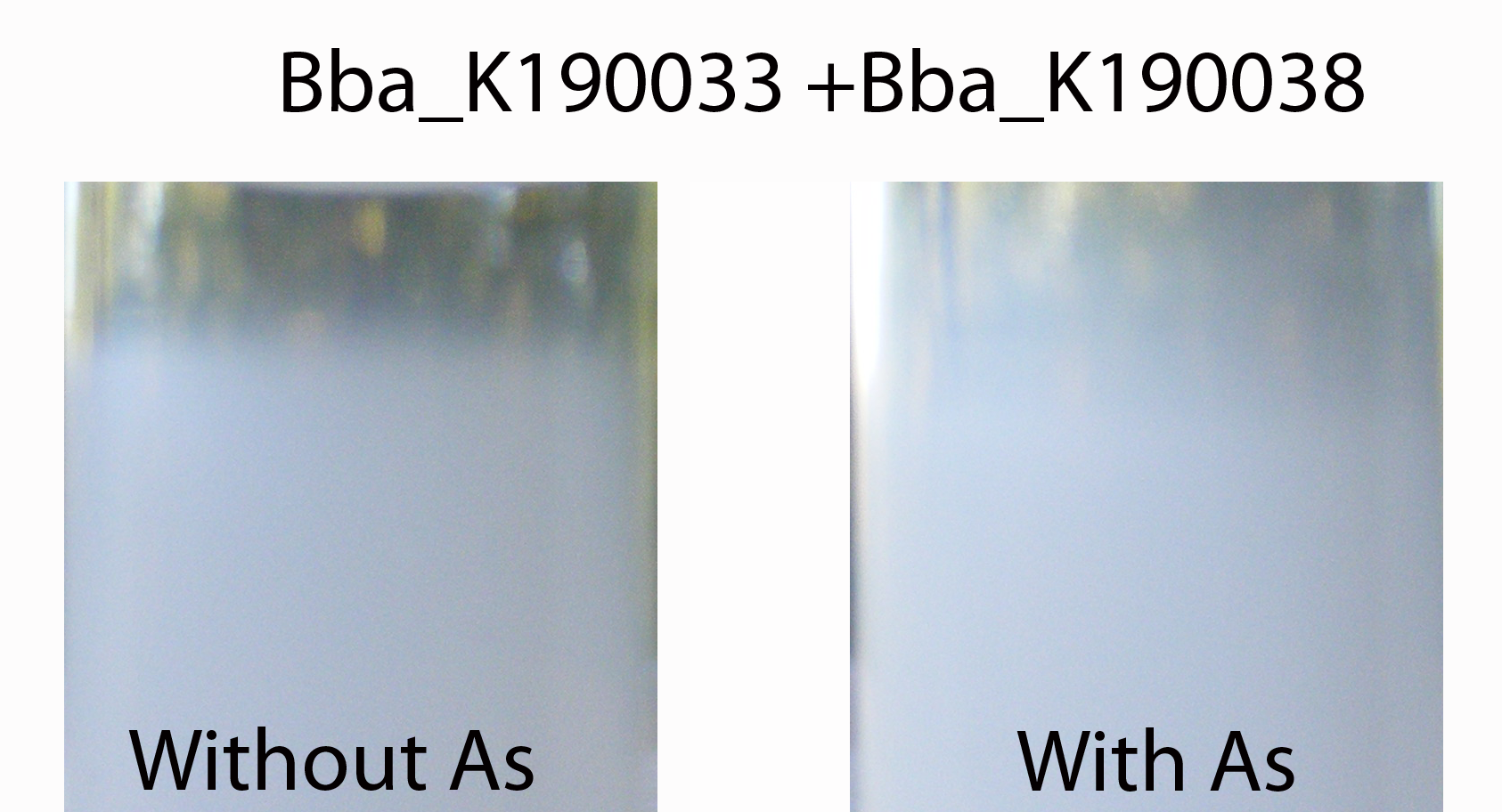
The buoyancy test elegantly shows that combining both an accumulation device and the gvp-buoyancy device allows buoyancy.
Figure 2
On the left E. coli cells (containing the whole system) without arsenic can be seen and on the right E. coli cells (containing the whole system) with arsenic is shown. It can be seen that the right sample floats. This indicates that the GlpF is transporting the arsenic inside the cells, the fMT accumulates it (otherwise the cells would be dead) and the gvp was induced by the arsenic so the cells start to float.
 "
"