Team:Heidelberg/k4l1um
From 2009.igem.org
(Difference between revisions)
(→6-17-2009) |
(→6-17-2009) |
||
| Line 56: | Line 56: | ||
* Annealing successful for JeT and Min | * Annealing successful for JeT and Min | ||
Annealing might not have been successful with Fos beacuse it contains a very repetetive proximal promoter. | Annealing might not have been successful with Fos beacuse it contains a very repetetive proximal promoter. | ||
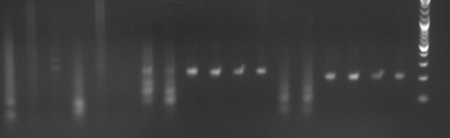
| - | [[Image:Bild1.jpg|frame|Lanes 1-6 Fos (403 Bp); Lanes 7-12 JeT (227 Bp): Lanes 13-18: Min (207 Bp) Lanes 1 : 1µL of 1 :10 diluted oligo 2 : 1 :100 3 : 1 :1000 ;4-6 same, but + DMSO Lanes 7,8,13,14: 1:10 Lanes 9,10,15,16: 1:100 Lanes 11,12,17,18: 1:1000 Even lanes: No DMSO Uneven lanes: DMSO| | + | [[Image:Bild1.jpg|frame|Lanes 1-6 Fos (403 Bp); Lanes 7-12 JeT (227 Bp): Lanes 13-18: Min (207 Bp) Lanes 1 : 1µL of 1 :10 diluted oligo 2 : 1 :100 3 : 1 :1000 ;4-6 same, but + DMSO Lanes 7,8,13,14: 1:10 Lanes 9,10,15,16: 1:100 Lanes 11,12,17,18: 1:1000 Even lanes: No DMSO Uneven lanes: DMSO|450px]] |
* gel purfifcation of JeT and Min | * gel purfifcation of JeT and Min | ||
* Repeat PCR of pcDNA5/FRT and mcherry with lower annealing temperature (58°C) and fresh Polymerase: Follow PCR protcol from Strategene site directed mutagenesis kit. | * Repeat PCR of pcDNA5/FRT and mcherry with lower annealing temperature (58°C) and fresh Polymerase: Follow PCR protcol from Strategene site directed mutagenesis kit. | ||
Revision as of 09:02, 24 June 2009
Contents |
6-15-2009
Lab: LV, SH, CZ
SH:
- Miniprep of GFP template plasmid, pcDNA5/FRT
| Nr | pcDNA5/FRT | GFP |
|---|---|---|
| 1 | 4,7 | 39,7 |
| 2 | 5,9 | 14,9 |
| 3 | 3,7 | 3,8 |
| 4 | 5,0 | 7,8 |
- Maxiprep pcDNA/FRT 188.7ng/µL
LV:
- Extraction of CMV promoter from 2008 distribution
- Transformation of DH5a cell with CMV promoter
- portzughtr
6-16-2009
Lab: LV, SH, CZ
- DNA synthesis (JeT, cFos, Min)
- Purification of cDNA via 2% / 3% agarosegel
=> Only bands at ca. 50 Bp => no successful amplification
- Site directed mutagenesis of pcDNA5/FRT and mcherry
- Dpn1 digestion
- Transformation of DH5a
6-17-2009
Lab: LV, SH
- No transformations could be observed
- Replace Phsuion stocks to Phusion Master Mix from Nathan
- Repeat DNA synthesis with a different protocol: 1:10 - 1:1000 dilution of Oligos, 95° 5', 7* [95° 45 58° 45 72° 1'], add 1:10 diluted primers, 95° 5' 25* same temperatures
- Annealing successful for JeT and Min
Annealing might not have been successful with Fos beacuse it contains a very repetetive proximal promoter.
- gel purfifcation of JeT and Min
- Repeat PCR of pcDNA5/FRT and mcherry with lower annealing temperature (58°C) and fresh Polymerase: Follow PCR protcol from Strategene site directed mutagenesis kit.
mcherry PCR might not have been successful because we used a template we didn't make ourselves - does it contain TE?
- PCR worked for pcDNA5FRT but not for mcherry
- DpnI digest of pcDNA5FRT 1,3,4,6
6-18-2009
Lab: LV, SH
- DNA synthesis for Fos (proximal) and Fos (core)
- Transformation of DH5a with pcDNA/FRT ΔPstI
Gave many 1000 colonies. Probably a contamination (?), whci hwould explain why there was no mutagenesis (see below)
- HeLa cells, MCS7 and U20S were splitted 1:3. Therefore DEMED medium (containing additionally 10% FCS, Pen-Strep, glutamate and non-essential aminoacids) was removed. Cells were washed afterwards with HBSS and trypsinised with 1 ml of trypsin and incubated for 5 min. at 37°C 5% CO2. The used flasks were filled up to a final volume of 7 ml with DMEM. 2 ml of the suspension was transfered to a new flask, 5 ml DMEM was added and incubated for another 3-4 days at 37°C and 5% CO2.
6-22-2009
Lab: LV, SH
- Split cells (Split again Thursday!)
- Prepare 6well-plate with U20S cells for Zeomycine assay
- Miniprep pcDNA/FRT ΔPstI
- Digest with PstI => No mutation
- Use Stratagene QuickChange XL kit for mutagenesis PCR (mcherry, pcDNAand Supercompetent Gold TOP10 cells for transformation => reasonable amount of colonies
6-23-2009
Lab: LV, SH
- Added Zeomycine to 6-well platees
- Miniprep mutagenisis PCR
- Digest with PstI => Mutagenesis successful
- PCR to remove EcoRI from pcDNA5/FRT using Phusion polymerase
6-24-2009
Lab: LV, SH
- DpnI digest
- Transformation of Supercompetent Gold TOP10 cells with pcDNA5 ΔEcoRI ΔPstI
6-25-2009
Split cells!
6-26-2009
Change 6well plate medium!
 "
"