Team:Heidelberg/stables
From 2009.igem.org
(→Results) |
(→LAM-PCR) |
||
| (16 intermediate revisions not shown) | |||
| Line 6: | Line 6: | ||
|-valign="top" border="0" style="margin-left: 2px;" | |-valign="top" border="0" style="margin-left: 2px;" | ||
|width="650px" style="padding: 0 15px 15px 20px; background-color:#ede8e2"| | |width="650px" style="padding: 0 15px 15px 20px; background-color:#ede8e2"| | ||
| - | + | =Stable Cell Line= | |
== Abstract == | == Abstract == | ||
| - | The one problem that is always faced when measuring any construct or device in mammalian cells is the fact that the transfection rate is not always constant. Thus, one never knows how many copies of a construct are actually in the cell. This is why we attempted to create a system that allows a controled integration of our constructs into the genome. To accomplish this we used the FRT/Flp system which is based on homologous recombination by the enzyme flippase at a specific sequence – the FRT site [[Team:Heidelberg/stables#References|[1]]]. One of those sequences is on a plasmid that is integrated randomly, but stably into the genome by antibiotic (Zeocin) selection. The other sequence is on the plasmid that contains the construct to be tested. Thus it would be possible to integrate at a specific site on the genome and select cells with the construct via a second antibiotic (Hygromycin). Additionally, the stable integration of an FRT site into the genome allows the characterization of the inserted constructs in the same genomic context thereby providing a normalization of the expression. Lastly, this integration site can also be located by LAM-PCR. | + | The one problem that is always faced when measuring any construct or device in mammalian cells is the fact that the transfection rate is not always constant. Thus, one never knows how many copies of a construct are actually in the cell. This is why we attempted to create a system that allows a controled integration of our constructs into the genome. To accomplish this we used the FRT/Flp system which is based on homologous recombination by the enzyme flippase at a specific sequence – the FRT site [[Team:Heidelberg/stables#References|[1]]]. One of those sequences is on a plasmid that is integrated randomly, but stably into the genome by antibiotic (Zeocin) selection. The other sequence is on the plasmid that contains the construct to be tested. Thus it would be possible to integrate at a specific site on the genome and select cells with the construct via a second antibiotic (Hygromycin). Additionally, the stable integration of an FRT site into the genome allows the characterization of the inserted constructs in the same genomic context thereby providing a normalization of the expression. Lastly, this integration site can also be located by [[Team:Heidelberg/stables#LAM-PCR|LAM-PCR]]. |
| - | == Introduction == | + | == Introduction== |
| - | + | [[Image:FRT.png|thumb|left|300px|<div style="text-align:justify;">'''Figure 1: Characteristics of FRT site.''' Cleavage and recombination by FLP takes place within the 8 bp spacer in between repeat 2 and 3. These two repeats are necessary for the reaction. Repeat 1 is present in most of the cases, but not strictly needed for the integration process.</div>]] | |
| - | [[Image:FRT.png|thumb|left|300px|<div style="text-align:justify;">''' | + | <div style="text-align:justify;">The FRT/Flp system originates from yeast. The eukaryotic site-specific recombinase FLP is originally encoded by the multicopy yeast plasmid 2µ circle DNA that is common to many yeast strains [[Team:Heidelberg/stables#References|[2]]][[Team:Heidelberg/stables#References|[3]]]. Naturally, the cleavage and recombination takes place at an 8 bp spacer that is on either side enclosed by two inverted repeats of 599 bp in length. For FLP-mediated recombination only 13 bp of each 599 bp repeat adjacent to the spacer and the spacer itself are necessary (see Fig. 1 sequences 2 and 3). Additionaly, most of the time another 13 bp repeat occurs immediately </div> [[Image:FRT_Flp.png|thumb|left|300px|<div style="text-align:justify;">'''Figure 2: Principle of FRT/Flp recombination.''' The integration of the GOI is realized by transfecting the cells with both the FRT plasmid containing the GOI and a plasmid which codes for the enzyme flippase. The integration process starting at the 8 bp spacer sequence of the FRT-sites occurs via Flip mediated homologous recombination. The process includes the destruction of the genomic Zeocin resistance and the integration of the Hygromycin resistance originally encoded on the plasmid, thus leading to new selection conditions for stable clones.</div>]] <div style="text-align:justify;">upstream of the 5’ repeat (see Fig. 1 sequence 1). The spacer contains the cleavage site and is thus very sensitive to alterations either in length or in sequence. Moreover, its asymmetry alone determines the orientation of the FLP recombination site [[Team:Heidelberg/stables#References|[2]]]. <br>Integrating one of those FLP recognition target sites (FRT site) stably into the genome and having another one on a plasmid which also contains the construct of interest will allow a controlled integration at that specific site by cotransfecting with an FLP encoding plasmid. Fig. 2 illustrates the nature of this integration. </div> |
| - | <div style="text-align:justify;">The FRT/Flp system originates from yeast. The eukaryotic site-specific recombinase FLP is originally encoded by the multicopy yeast plasmid 2µ circle DNA that is common to many yeast strains [[Team:Heidelberg/stables#References|[2]]][[Team:Heidelberg/stables#References|[3]]]. Naturally, the cleavage and recombination takes place at an 8 bp spacer that is on either side enclosed by two inverted repeats of 599 bp in length. For FLP-mediated recombination only 13 bp of each 599 bp repeat adjacent to the spacer and the spacer itself are | + | |
<br><br><br><br><br><br><br> | <br><br><br><br><br><br><br> | ||
<br> | <br> | ||
| Line 20: | Line 19: | ||
== Results == | == Results == | ||
| - | We attempted to make a cell line with an FRT site by using the pFRT/''lacZeo'' plasmid (Invitrogen). After transfecting HeLa, U2-OS and MCF-7 cells with the mentioned pFRT/''lacZeo'' plasmid by using the Effectene kit (Qiagen) we selected cells that stably integrated the plasmid by adding Zeocin (100 μg/ml) to the culture medium (DMEM+++). After several days, cells with a Zeocin resistance | + | We attempted to make a cell line with an FRT site by using the [[Team:Heidelberg/Notebook_MaM#Plasmids|pFRT/''lacZeo'' plasmid]] (Invitrogen). After transfecting HeLa, U2-OS and MCF-7 cells with the mentioned pFRT/''lacZeo'' plasmid by using the [[Team:Heidelberg/Notebook_MaM#Transfection_of_Mammalian_cells|Effectene kit]] (Qiagen), we selected cells that stably integrated the plasmid by adding Zeocin (100 μg/ml) to the culture medium ([[Team:Heidelberg/Notebook_MaM|DMEM+++]]). After several days, cells with a Zeocin resistance survive and grow in foci originated from one single cell. Those foci were then [[Team:Heidelberg/Notebook_MaM#Picking_cells_with_cloning_disks|picked]] as described in the cell culture methods. They were expanded into culture flasks and aliquots were frozen. After DNA extraction by use of the [[Team:Heidelberg/Notebook_MaM#Kits|High Pure PCR Template Preparation Kit]] (Roche), [[Team:Heidelberg/stables#LAM-PCR|LAM-PCR]] was carried out to localize the integration sites on the genome. Cotransfection of the plasmids [[Team:Heidelberg/Notebook_MaM#Plasmids|pOG44]] (encodes for FLP) and [[Team:Heidelberg/Notebook_MaM#Plasmids|p55]] (contains CMV promoter and GFP) at a ratio of 9:1 led to the integration of a GFP with a CMV promoter in some of the cells. After this cotransfection the cells were kept under Hygromycin selection (200 µg/ml). There is no promoter in front of the resistance gene for Hygromycin so that cells that randomly and stably integrate the plasmid (as it was done for the pFRT/''lacZeo'') do not express the resistance. Once again, foci were visible after several days( see Fig. 3). |
| - | To properly characterize the integration sites of the pFRT/''lacZeo'' plasmid in the genome we performed LAM-PCR at Thereby we confirmed | + | To properly characterize the integration sites of the pFRT/''lacZeo'' plasmid in the genome we performed [[Team:Heidelberg/stables#LAM-PCR|LAM-PCR]] at M. Schmidt's lab (see [[Team:Heidelberg/stables#References|reference 5]] for details) at the nct (national center for tumor diseases) in Heidelberg. Thereby we confirmed that we were able to generate HeLa, MCF-7 and U2-OS cells that stably integrated the FRT-site into their genome. For each cell line there are several bands visible (see Fig. 4). Since the number of bands correlates with the number of unique integration sites [[Team:Heidelberg/stables#References|[5]]] there must have been more than one integration of the FRT-vector into the genome of the cells. |
{| | {| | ||
|-valign="top" border="0" | |-valign="top" border="0" | ||
| | | | ||
| - | [[Image:Stablegreen1.png|thumb|left|300px|<div style="text-align:justify;">''' | + | [[Image:Stablegreen1.png|thumb|left|300px|<div style="text-align:justify;">''' Figure 3: One focus of cells''' that stably integrated a plasmid containing CMV and GFP into their genome. Cells in the focus should all have evolved from a single cell, that successfully passed through homologous recombination at the FRT- site mediated by flippase </div>]] |
| | | | ||
| - | [[Image:Gel_LAM-PCR1.png|thumb|left|300px|<div style="text-align:justify;">''' | + | [[Image:Gel_LAM-PCR1.png|thumb|left|300px|<div style="text-align:justify;">'''Figure 4: Results of second exponential PCR.''' In all lanes there are several bands visible, indicating that there is more than one FRT-site integrated in each cell line. Negative controls are: 1. untransfected genomic DNA 2. H<sub>2</sub>O negative control LAM-PCR, 3. H<sub>2</sub>O negative control first exponential PCR, 4. H<sub>2</sub>O negative control second exponential PCR. Electrophoresis was carried out on 2% agarose; [http://tools.invitrogen.com/content/sfs/manuals/15628019.pdf 100 bp DNA ladder] was used.</div>]] |
|- | |- | ||
|} | |} | ||
== Discussion == | == Discussion == | ||
| + | |||
| + | Our experiment showed that it is possible to generate cell lines with stable FRT sites. We are aware that our cell lines with multiple FRT sites are not ideal for the characterization of promoters. Nevertheless, we are convinced that a cell line that contains a single integration site can be generated and that it would be very valuable for standardized characterization of BioBrick parts in eukaryotes. Even though those characterizations are more time consuming due to the Hygromycin selection, they are of advantage once established, because the construct cannot be "washed out" as it is observed when dealing with transiently transfected cells. | ||
| + | Measurements with our cell lines with multiple integration sites are also possible and favorable over transiently transfected cell lines. For one, it is expected that the number of construct integrations is statistically distributed. Furthermore, one can determine the number of constructs integrated. Since the genomic surroundings of the integration sites can be characterized by [[Team:Heidelberg/stables#LAM-PCR|LAM-PCR]], another (regular) PCR with one primer that is located on the genome and one that is on the other side of the gene of interest (GOI) can be carried out. | ||
| + | To generate a cell line with only one integration site, the pFRT/''lacZeo'' should have been linearized before transfection. That way, the transfection efficiency and thus the chance to stably integrate more than one copy would have been lower. Also, linearization of the plasmid is favorable for LAM-PCR, because the sequences directly adjacent to the genomic DNA would be known. Although being very time-consuming, another attempt to create a cell line that stably integrated only one FRT-site would have a great impact for the synthetic biology community. | ||
== Detailed Methods == | == Detailed Methods == | ||
=== LAM-PCR === | === LAM-PCR === | ||
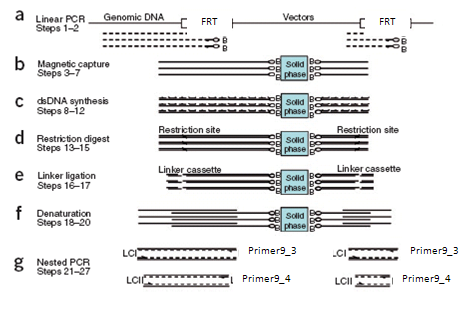
| - | + | [[Image:LAM.png|thumb|left|350px|<div style="text-align:justify;">'''Figure 5: Schematic outline of LAM-PCR used to amplify the genomic DNA flanking the FRT-site.'''The solid phase consists of Streptavidin-coupled magnetic beads which allow for the binding of DNA via biotinylated Primers.</div>]] <div style="text-align:justify;"> In order to determine where the plasmid containing the FRT-site is integrated into the genome, LAM-PCR is applied. This method established by Schmidt ''et al''. can be used for identifying all unknown DNA sequences adjacent to known DNA-sequences.Based on linear amplification-mediated PCR, it allows for amplifying the sequences flanking the integrated vector thus supplying the information which is needed for alignment with the host genome. | |
| - | [[Image:LAM.png|thumb|left|350px|<div style="text-align:justify;">''' | + | |
The procedure starts with linear PCR (Fig 5.a), which is carried out twice with 50 - cycles for each reaction. Primers used in PCR are labeled with biotin, which has a high affinity to Streptavidin [[Team:Heidelberg/stables#References|[4]]]. By coupling Streptavidin to magnetic beats, removal of non-target DNA can be achieved via magnetic selection: newly synthesized single strands will bind via primer to the magnetic beads and can then be captured with MPC (Magnetic particle concentrator). The same kind of selection is used for the subsequent steps. | The procedure starts with linear PCR (Fig 5.a), which is carried out twice with 50 - cycles for each reaction. Primers used in PCR are labeled with biotin, which has a high affinity to Streptavidin [[Team:Heidelberg/stables#References|[4]]]. By coupling Streptavidin to magnetic beats, removal of non-target DNA can be achieved via magnetic selection: newly synthesized single strands will bind via primer to the magnetic beads and can then be captured with MPC (Magnetic particle concentrator). The same kind of selection is used for the subsequent steps. | ||
| - | After linear amplification complementary double strands are synthesized by hexanucleotide priming: a hexanucleotide mixture, dNTPS and Polymerase are added to the single stranded PCR-products. Hexanucleotides will prime to the single stranded DNA and polymerase will fill the gaps in between (Fig. 5.c) | + | After linear amplification, complementary double strands are synthesized by hexanucleotide priming: a hexanucleotide mixture, dNTPS and Polymerase are added to the single stranded PCR-products. Hexanucleotides will prime to the single stranded DNA and polymerase will fill the gaps in between (Fig. 5.c) |
Those double strands are cut with restriction enzymes and a linker cassette with known sequence is ligated to the genomic end of the fragments which sequences are unknown (Fig. 5.d,e). | Those double strands are cut with restriction enzymes and a linker cassette with known sequence is ligated to the genomic end of the fragments which sequences are unknown (Fig. 5.d,e). | ||
This step is followed by denaturation of the double stranded DNA composed of linker cassette, genomic DNA and vector DNA by sodium hydroxid. The resulting single stranded DNA is then amplified by exponential PCR, using primers that prime to linker cassette and vector DNA. Another denaturation and a second exponential PCR is carried out (Fig. 5.f,g). | This step is followed by denaturation of the double stranded DNA composed of linker cassette, genomic DNA and vector DNA by sodium hydroxid. The resulting single stranded DNA is then amplified by exponential PCR, using primers that prime to linker cassette and vector DNA. Another denaturation and a second exponential PCR is carried out (Fig. 5.f,g). | ||
Latest revision as of 21:51, 21 October 2009
Stable Cell LineAbstractThe one problem that is always faced when measuring any construct or device in mammalian cells is the fact that the transfection rate is not always constant. Thus, one never knows how many copies of a construct are actually in the cell. This is why we attempted to create a system that allows a controled integration of our constructs into the genome. To accomplish this we used the FRT/Flp system which is based on homologous recombination by the enzyme flippase at a specific sequence – the FRT site [1]. One of those sequences is on a plasmid that is integrated randomly, but stably into the genome by antibiotic (Zeocin) selection. The other sequence is on the plasmid that contains the construct to be tested. Thus it would be possible to integrate at a specific site on the genome and select cells with the construct via a second antibiotic (Hygromycin). Additionally, the stable integration of an FRT site into the genome allows the characterization of the inserted constructs in the same genomic context thereby providing a normalization of the expression. Lastly, this integration site can also be located by LAM-PCR. IntroductionThe FRT/Flp system originates from yeast. The eukaryotic site-specific recombinase FLP is originally encoded by the multicopy yeast plasmid 2µ circle DNA that is common to many yeast strains [2][3]. Naturally, the cleavage and recombination takes place at an 8 bp spacer that is on either side enclosed by two inverted repeats of 599 bp in length. For FLP-mediated recombination only 13 bp of each 599 bp repeat adjacent to the spacer and the spacer itself are necessary (see Fig. 1 sequences 2 and 3). Additionaly, most of the time another 13 bp repeat occurs immediately 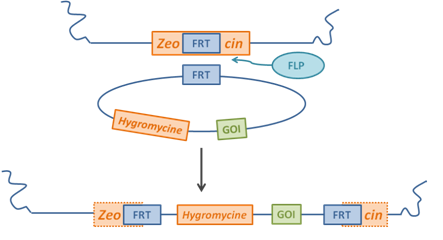 Figure 2: Principle of FRT/Flp recombination. The integration of the GOI is realized by transfecting the cells with both the FRT plasmid containing the GOI and a plasmid which codes for the enzyme flippase. The integration process starting at the 8 bp spacer sequence of the FRT-sites occurs via Flip mediated homologous recombination. The process includes the destruction of the genomic Zeocin resistance and the integration of the Hygromycin resistance originally encoded on the plasmid, thus leading to new selection conditions for stable clones. upstream of the 5’ repeat (see Fig. 1 sequence 1). The spacer contains the cleavage site and is thus very sensitive to alterations either in length or in sequence. Moreover, its asymmetry alone determines the orientation of the FLP recombination site [2].
Integrating one of those FLP recognition target sites (FRT site) stably into the genome and having another one on a plasmid which also contains the construct of interest will allow a controlled integration at that specific site by cotransfecting with an FLP encoding plasmid. Fig. 2 illustrates the nature of this integration.
ResultsWe attempted to make a cell line with an FRT site by using the pFRT/lacZeo plasmid (Invitrogen). After transfecting HeLa, U2-OS and MCF-7 cells with the mentioned pFRT/lacZeo plasmid by using the Effectene kit (Qiagen), we selected cells that stably integrated the plasmid by adding Zeocin (100 μg/ml) to the culture medium (DMEM+++). After several days, cells with a Zeocin resistance survive and grow in foci originated from one single cell. Those foci were then picked as described in the cell culture methods. They were expanded into culture flasks and aliquots were frozen. After DNA extraction by use of the High Pure PCR Template Preparation Kit (Roche), LAM-PCR was carried out to localize the integration sites on the genome. Cotransfection of the plasmids pOG44 (encodes for FLP) and p55 (contains CMV promoter and GFP) at a ratio of 9:1 led to the integration of a GFP with a CMV promoter in some of the cells. After this cotransfection the cells were kept under Hygromycin selection (200 µg/ml). There is no promoter in front of the resistance gene for Hygromycin so that cells that randomly and stably integrate the plasmid (as it was done for the pFRT/lacZeo) do not express the resistance. Once again, foci were visible after several days( see Fig. 3). To properly characterize the integration sites of the pFRT/lacZeo plasmid in the genome we performed LAM-PCR at M. Schmidt's lab (see reference 5 for details) at the nct (national center for tumor diseases) in Heidelberg. Thereby we confirmed that we were able to generate HeLa, MCF-7 and U2-OS cells that stably integrated the FRT-site into their genome. For each cell line there are several bands visible (see Fig. 4). Since the number of bands correlates with the number of unique integration sites [5] there must have been more than one integration of the FRT-vector into the genome of the cells. DiscussionOur experiment showed that it is possible to generate cell lines with stable FRT sites. We are aware that our cell lines with multiple FRT sites are not ideal for the characterization of promoters. Nevertheless, we are convinced that a cell line that contains a single integration site can be generated and that it would be very valuable for standardized characterization of BioBrick parts in eukaryotes. Even though those characterizations are more time consuming due to the Hygromycin selection, they are of advantage once established, because the construct cannot be "washed out" as it is observed when dealing with transiently transfected cells. Measurements with our cell lines with multiple integration sites are also possible and favorable over transiently transfected cell lines. For one, it is expected that the number of construct integrations is statistically distributed. Furthermore, one can determine the number of constructs integrated. Since the genomic surroundings of the integration sites can be characterized by LAM-PCR, another (regular) PCR with one primer that is located on the genome and one that is on the other side of the gene of interest (GOI) can be carried out. To generate a cell line with only one integration site, the pFRT/lacZeo should have been linearized before transfection. That way, the transfection efficiency and thus the chance to stably integrate more than one copy would have been lower. Also, linearization of the plasmid is favorable for LAM-PCR, because the sequences directly adjacent to the genomic DNA would be known. Although being very time-consuming, another attempt to create a cell line that stably integrated only one FRT-site would have a great impact for the synthetic biology community. Detailed MethodsLAM-PCR In order to determine where the plasmid containing the FRT-site is integrated into the genome, LAM-PCR is applied. This method established by Schmidt et al. can be used for identifying all unknown DNA sequences adjacent to known DNA-sequences.Based on linear amplification-mediated PCR, it allows for amplifying the sequences flanking the integrated vector thus supplying the information which is needed for alignment with the host genome.
The procedure starts with linear PCR (Fig 5.a), which is carried out twice with 50 - cycles for each reaction. Primers used in PCR are labeled with biotin, which has a high affinity to Streptavidin [4]. By coupling Streptavidin to magnetic beats, removal of non-target DNA can be achieved via magnetic selection: newly synthesized single strands will bind via primer to the magnetic beads and can then be captured with MPC (Magnetic particle concentrator). The same kind of selection is used for the subsequent steps. After linear amplification, complementary double strands are synthesized by hexanucleotide priming: a hexanucleotide mixture, dNTPS and Polymerase are added to the single stranded PCR-products. Hexanucleotides will prime to the single stranded DNA and polymerase will fill the gaps in between (Fig. 5.c) Those double strands are cut with restriction enzymes and a linker cassette with known sequence is ligated to the genomic end of the fragments which sequences are unknown (Fig. 5.d,e). This step is followed by denaturation of the double stranded DNA composed of linker cassette, genomic DNA and vector DNA by sodium hydroxid. The resulting single stranded DNA is then amplified by exponential PCR, using primers that prime to linker cassette and vector DNA. Another denaturation and a second exponential PCR is carried out (Fig. 5.f,g). PCR-products are visualized by separation on a 2% agarose gel. For a detailed description see [5]References[1] Zhou H. Development of site-specific integration system to high-level expression recombinant proteins in CHO cells. Chinese journal of biotechnology 23: 756-62 (2007). [2] Andrews B. J., Proteau G. A., Beatty L. G. & Sadowski P. D. The FLP Recombinase of the 2μ Circle DNA of Yeast: Interaction with Its Target Sequences. Cell 40: 795-803 (1985). [3] Senecoff J. F., Bruckner R. C. & Cox M. M. The FLP recombinase of the yeast 2-μ plasmid: Characterization of its recombination site. Proc. Nati. Acad. Sci. USA 82: 7270-7274 (1985). [4] Holmberg A., Blomstergren A., Nord O., Lukacs M., Lundeberg J. & Uhlén M. The biotin-streptavidin interaction can be reversibly broken using water at elevated temperatures. Electrophoresis 26(3): 501-10 (2005). [5] Schmidt M., Schwarzwaelder K., Bartholomae C., Zaoui K., Ball B.,Pilz I.,Braun S.,Glimm H.,von Kalle C., High-resolution insertion-site analysis by linear amplification–mediated PCR (LAM-PCR). Nature Methods 4(12): 1051-1057 (2005).
|
 "
"