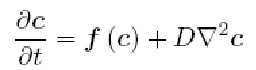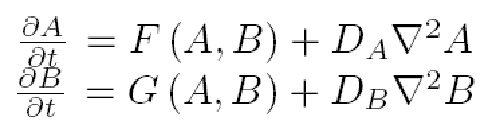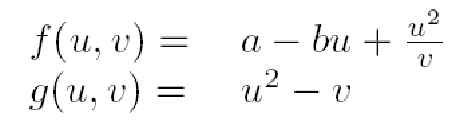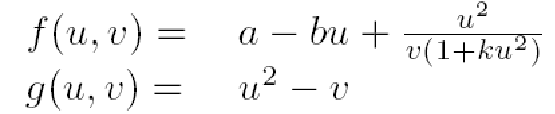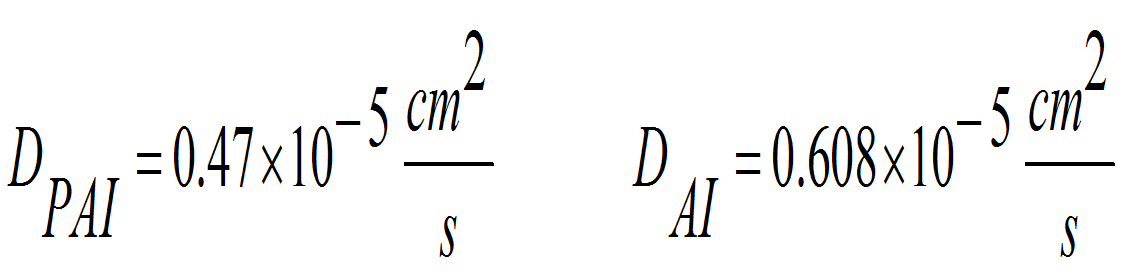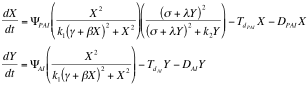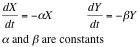Team:IPN-UNAM-Mexico/Modeling
From 2009.igem.org
Modelling
 Introduction: Developing an Activator-Inhibitor network
Introduction: Developing an Activator-Inhibitor network
 Introduction: Developing an Activator-Inhibitor network
Introduction: Developing an Activator-Inhibitor network
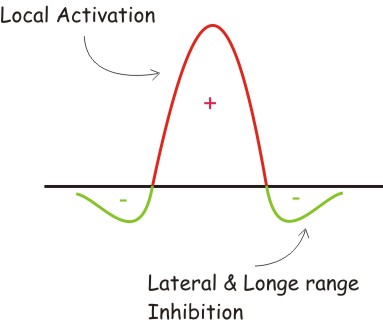
As described on the overview, the spatiotemporal patterns we are looking forward to reproduce on the lab were based on the equations of reaction-diffusion with a chemical kinetics' and diffusive behavior of morphogens in an activator-inhibitor system. The qualitative dynamics of this kind of systems such as local activation and long range inhibition were the base and theoretical framework to generate our BioBricks synthetic network.
The following equation is the general expression of a reaction-diffusion system, which can be seen as a generalization of the [http://en.wikipedia.org/wiki/Heat_equation heat equation]. This tells us broadly speaking that the change on time of the reactants concentration depends on their mutual interaction and their diffusion over the space; this last term being defined by coefficients of D (the diagonal matrix with the diffusion constants or the reactants) times the Laplace operator.
We will represent the dynamics of our network as an activator-inhibitor system, so we will need only a few equations to describe it, where F and G are the functions that describe the chemical kinetics of morphogens A and B, with diffusion coefficient constants D_A and D_B:
Gierer and Meinhardt proposed in 1972 a system of reaction diffusion equations that fulfill the Turing hypothesis for generating spatiotemporal patterns. In this particular systems A stands for the activator and B for the inhibitor in:
Which with the proper adimentionalizing analysis leads to:
and including the activator's inhibition results in:
where a and b are positive constants and k is a measure of the strength of the inhibition.
As introduced in the project section and full described in the parts section we designed a synthetic network with the qualitative properties of an activator-inhibitor system.
Along with the developing of the project we continued to gather the information required to model the activator-inhibitor system as an approximation of the BioBricks network. We also needed the diffusion constants of PAI and AI, since its chemical nature makes a big difference for the formation of the patterns.
 First approach: Classical model with estimated diffusion constants
First approach: Classical model with estimated diffusion constants
 First approach: Classical model with estimated diffusion constants
First approach: Classical model with estimated diffusion constants
To do this we considered the molecular weight of the AHL molecules and we estimated the diameter of the molecules taking into account the linear structure of them. From the articles of A. Einstein: “On the Motion—Required by the Molecular Kinetic Theory of Heat—of Small Particles Suspended in a Stationary Liquid” and “A new determination of molecular dimensions” we used the method to estimate the diffusion coefficients from the diameter of the molecule using the modified friction coefficient of the water in presence of glucose to improve the accuracy as described.
The equations are:
Obtaining the following values:
Using Comsol Multipysics [http://www.comsol.com/ Comsol Multipysics] we made a simulation with values previously used by Murray [14] and the previously estimated diffusion coefficients of PAI and AI molecules with the reaction-diffusion equations of the Gierer and Meinhardt model. The simulation was made in a circular area with zero flux boundary conditions and all the parameters values found in the literature.
As expected we obtained a spots pattern in a specific diameter value and a stripes pattern using a smaller diameter, both starting with a sinusoidal distribution of the activator and the inhibitor.
 Second approach: Activator-Inhibitor dynamics on a single cell
Second approach: Activator-Inhibitor dynamics on a single cell
 Second approach: Activator-Inhibitor dynamics on a single cell
Second approach: Activator-Inhibitor dynamics on a single cell
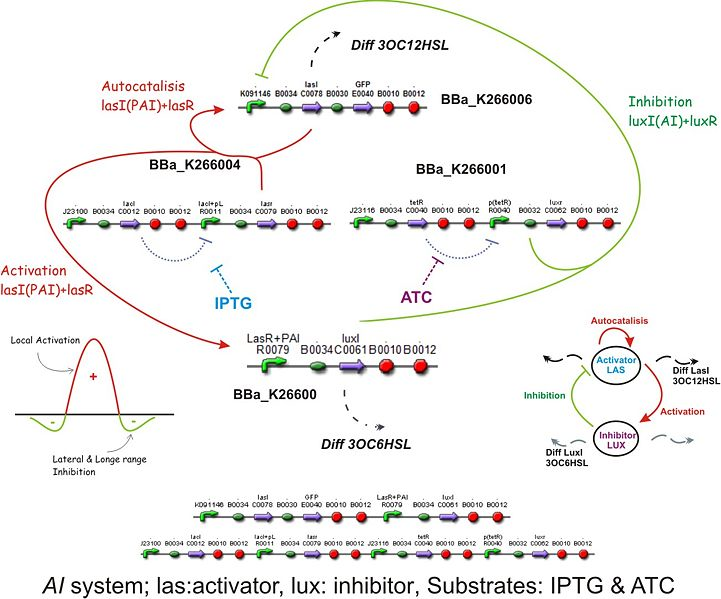
Based on the design of the biobricks network we tried to model the behavior it would have within a single cell. To do this we constructed the kinetic laws describing the relationships between the components of our network and IPTG and aTc. We modeled the interactions of the AHLs with their respective targets as allosteric regulators affecting the production rate of the catalyzing enzymes, thus affecting the synthesis of the AHLs. This catalysis was assumed to be a first order kinetics depending only on the concentration of the catalyst.
The diffusion rate of the AHLs was reduced to a first order kinetics considering the membrane area of the bacteria and assuming the complete absence of the AHL outside the cell, therefore depending only on the production within the cell.
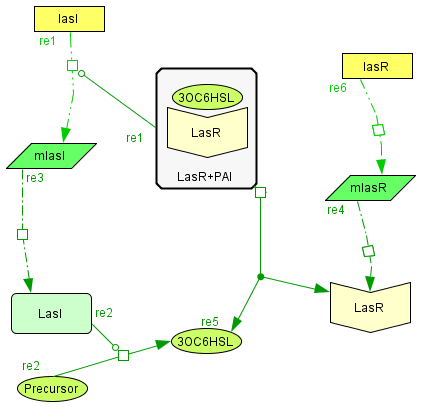
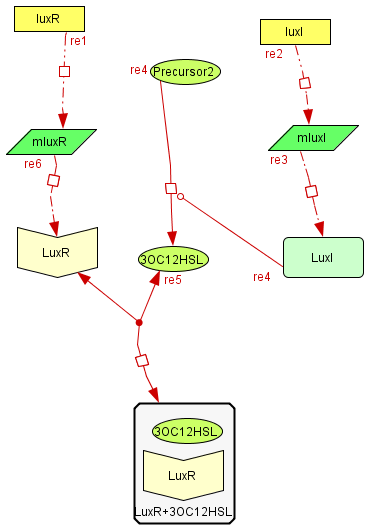
The Activator module
| The activator module is a representation of the kinetiks involved in the same module on the BioBricks design. In this module the transcription and translation of the gene lasR to the protein LasR induces the association with its activator, wich cathalyzes the transcriprion of las. The increasing of its translation finally leads to an increase of the cathalytic production of the activator. |
The Inhibitory module
| The inhibitory module consists on two separated genes luxI and luxR producing ther respective proteins. LuxI is the cathalizer of the Lux operon activator, and LuxR is the receptor for this activator. The way this system inhibits is by the binding of the complex LuxR and its activator to a promoter repressible by this complex. |
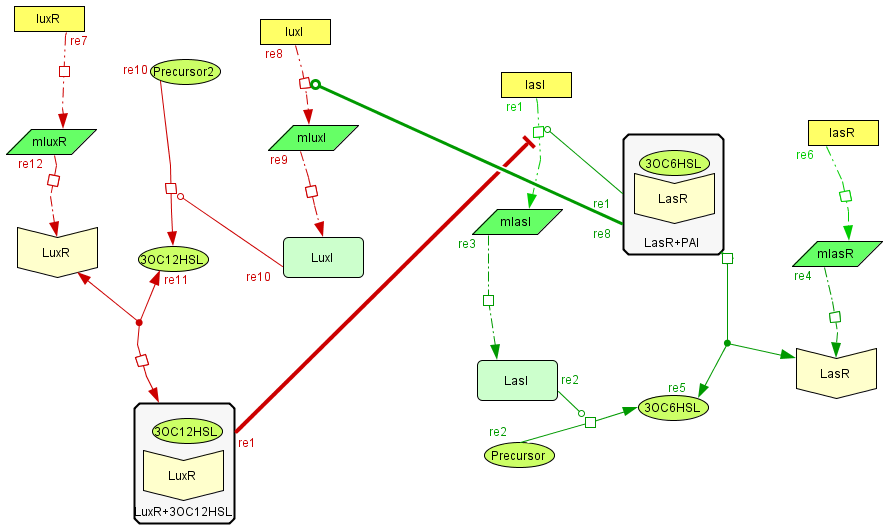
Activator-Inhibitor interaction
| The interaction between the activator and the inhibitor is given by the inhibition on the transcription of lasI and by the induction on the transcription of the luxI gene. This interaction qualitatively behaves as the theoretical Activator-Inhibitor system. |
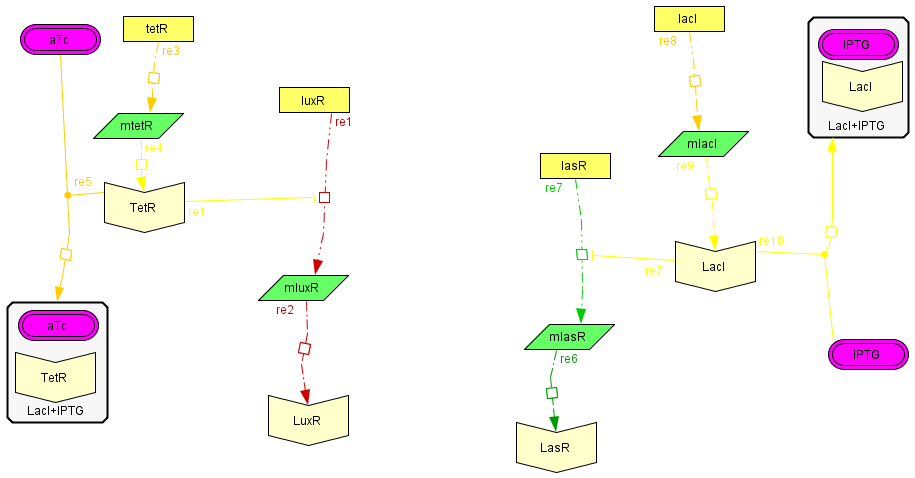
The regulatory modules
| To introduce the regulatory modules on this model we explicitely described the mechanistic description of the Lac and Tet inversor coupled to their regulatory targets: lasR and luxR, respectively;wich are essential components of the activator-inhibytor system. In this way when we add IPTG or aTc the inhibition on the transcription of the genes is removed, increasing the production of the proteins. We also observed that the concentration of the complex (LacI+IPTG or Tet+aTc) eventually reaches to a steady state, allowing us to consider this variables as constant in the following model. The amount of free inhibitor varies depending on the amount of IPTG or aTc we add, so we can regulate the degree of inhibition acting on the activator and inhibitory module |
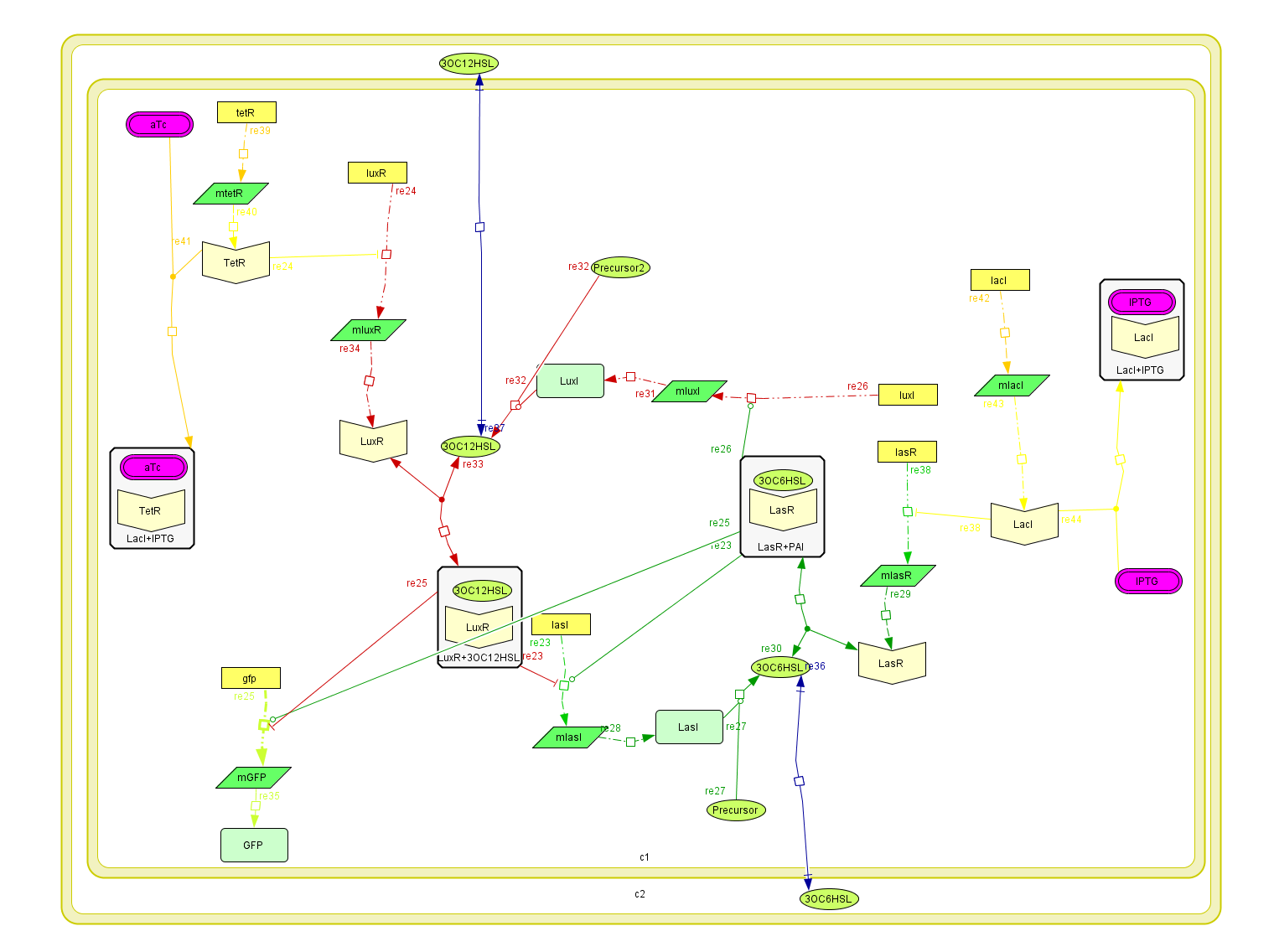
The integrated system
To build up the entire model we put together all the previous submodels, adding the diffusion of the AHLs and introducing a reporter moduel under the same regulation as lasI.
This system was built on [http://www.celldesigner.org/ Cell Designer ] software to explicitly denoting all the described interactions. Each coefficient of the kinetic rules was gathered from the literature and past years [http://parts.mit.edu/igem07/index.php?title=ETHZ team wikis].
For this model. We tried to be very accurate on the description of our system, and the parameters and the kinetic laws we used are described below.
| Constant | Value | Original Units |
| Kc PAI | 0.45
| |
| G PAI | 300
| copies |
| Tmax PAI | 0.01
| mM/h |
| TR PAI | 0.556
| 1/s |
| Td PAI | 0.007
| per protein |
| D PAI | 4.30E-06
| cm2/s |
| kd1 (PAI) | 250
| nM |
| kd2 (AI) | 250
| nM |
| kPAI | 0.5
| uM |
| Kc LasR | 0.45
| |
| G LasR | 300
| copies |
| Tmax LasR | 0.01
| mM/h |
| TR LasR | 0.556
| 1/s |
| kd LacI | 800
| nM |
| kAI | 0.5
| uM |
| Kc LuxR | 0.45
| |
| G LuxR | 300
| copies |
| Tmax LuxR | 0.01
| mM/h |
| TR LuxR | 0.556
| 1/s |
| kd TetR | 179
| pM |
| Kc AI | 0.45
| |
| G AI | 300
| copies |
| Tmax AI | 0.01
| mM/h |
| TR AI | 0.556
| 1/s |
| Td AI | 0.007
| per protein |
| D AI | 6.00E-06
| cm2/s |
| [Iptg] | 1
| mM |
| [aTc] | 3
| uM |
| [LacI] inicial T | 0.008
| mM |
| [TetR] inicial T | 0.008
| mM |
| kd iptg lacI | 893
| pM |
| kd aTc tetR | 1.3
| uM |
 Third approach: Spatial model
Third approach: Spatial model
 Third approach: Spatial model
Third approach: Spatial model
Afterward we described as ordinary differential equations the time change of the concentrations of PAI and AI based on the kinetic law equations, being x PAI and y AI
Here all the constants are sums or products of kinetic constants.
With this system of differential equations describing the kinetic basis of the reaction and diffusion we proceeded to make a qualitative analysis of its behavior as a 2x2 system of equations using [http://en.wikipedia.org/wiki/Grapher Grapher].
From this analysis we can see simply by looking at its vectorial field that it behaves as the system:
We wanted to check if this equations as chemical kinetics into a system of the reaction-diffusion dynamics can produce spatiotemporal patterns; to do this we made a simulation using [http://www.comsol.com/ ComsolMultipysics] as previously described
 References
References
 References
References
[1] Weber W et al. "A synthetic time-delay circuit in mammalian cells and mice", P Natl Acad Sci USA 104(8):2643-2648, 2007.
[2] Setty Y et al. "Detailed map of a cis-regulatory input function", P Natl Acad Sci USA 100(13):7702-7707, 2003.
[3] Braun D et al. "Parameter Estimation for Two Synthetic Gene Networks: A Case Study", ICASSP 5:769-772, 2005.
[4] Fung E et al. "A synthetic gene--metabolic oscillator", Nature 435:118-122, 2005 (supplementary material).
[5] Iadevaia S and Mantzais NV "Genetic network driven control of PHBV copolymer composition", J Biotechnol 122(1):99-121, 2006.
[6] Goryachev AB et al. "Systems analysis of a quorum sensing network: Design constraints imposed by the functional requirements, network topology and kinetic constants", Biosystems 83(2-3):178-187, 2004.
[7] Arkin A et al. "Stochastic kinetic analysis of developmental pathway bifurcation in phage λ-Infected Escherichia coli cells", Genetics 149: 1633-1648, 1998.
[8] Colman-Lerner A et al. "Yeast Cbk1 and Mob2 Activate Daughter-Specific Genetic Programs to Induce Asymmetric Cell Fates", Cell 107(6): 739-750, 2001 (supplementary material).
[9] Becskei A and Serrano L "Engineering stability in gene networks by autoregulation", Nature 405: 590-593, 2000.
[10] Tuttle et al. "Model-Driven Designs of an Oscillating Gene Network", Biophys J 89(6):3873-3883, 2005.
[11] McMillen LM et al. "Synchronizing genetic relaxation oscillators by intercell signaling", P Natl Acad Sci USA 99(2):679-684, 2002.
[12] Basu S et al. "A synthetic multicellular system for programmed pattern formation", Nature 434:1130-1134, 2005.
[13] Einstein, Albert , "On the Motion—Required by the Molecular Kinetic Theory of Heat—of Small Particles Suspended in a Stationary Liquid", 1905.
[14] Einstein, Albert , "A new determination of molecular dimensions", 1905.
[15] Gierer A and Meinhardt H, "A theory of biologycal pattern formation", Kybernetik 12:30-39, 1972
[16] Turing AM, "The Chemical Basis of Morphogenesis", Philosophical Transactions of The Royal Society of London, series B, 237:37–72, 1952.
[17] Wolpert L, "Positional information and the spatial pattern of cellular differentiation", Journal of Theoretical Biology, 25:1–47, 1969
[18] Murray JD, "Mathematical Biology II", Springer-Verlag Berlin Heidelberg, 3rd edition, 1993





 "
"