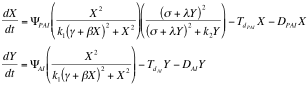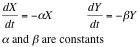Team:IPN-UNAM-Mexico/Modeling
From 2009.igem.org
Modelling
 Introduction: Developing an Activator-Inhibitor network
Introduction: Developing an Activator-Inhibitor network
 First approach: Classical model with estimated diffusion constants
First approach: Classical model with estimated diffusion constants
 Second approach: Activator-Inhibitor dynamics on a single cell
Second approach: Activator-Inhibitor dynamics on a single cell
 Third approach: Spatial model
Third approach: Spatial model
 Third approach: Spatial model
Third approach: Spatial model
Afterward we described as ordinary differential equations the time change of the concentrations of PAI and AI based on the kinetic law equations, being x PAI and y AI
Here all the constants are sums or products of kinetic constants.
With this system of differential equations describing the kinetic basis of the reaction and diffusion we proceeded to make a qualitative analysis of its behavior as a 2x2 system of equations using [http://en.wikipedia.org/wiki/Grapher Grapher].
From this analysis we can see simply by looking at its vectorial field that it behaves as the system:
We wanted to check if this equations as chemical kinetics into a system of the reaction-diffusion dynamics can produce spatiotemporal patterns; to do this we made a simulation using [http://www.comsol.com/ ComsolMultipysics] as previously described
 References
References
 References
References
[1] Weber W et al. "A synthetic time-delay circuit in mammalian cells and mice", P Natl Acad Sci USA 104(8):2643-2648, 2007.
[2] Setty Y et al. "Detailed map of a cis-regulatory input function", P Natl Acad Sci USA 100(13):7702-7707, 2003.
[3] Braun D et al. "Parameter Estimation for Two Synthetic Gene Networks: A Case Study", ICASSP 5:769-772, 2005.
[4] Fung E et al. "A synthetic gene--metabolic oscillator", Nature 435:118-122, 2005 (supplementary material).
[5] Iadevaia S and Mantzais NV "Genetic network driven control of PHBV copolymer composition", J Biotechnol 122(1):99-121, 2006.
[6] Goryachev AB et al. "Systems analysis of a quorum sensing network: Design constraints imposed by the functional requirements, network topology and kinetic constants", Biosystems 83(2-3):178-187, 2004.
[7] Arkin A et al. "Stochastic kinetic analysis of developmental pathway bifurcation in phage λ-Infected Escherichia coli cells", Genetics 149: 1633-1648, 1998.
[8] Colman-Lerner A et al. "Yeast Cbk1 and Mob2 Activate Daughter-Specific Genetic Programs to Induce Asymmetric Cell Fates", Cell 107(6): 739-750, 2001 (supplementary material).
[9] Becskei A and Serrano L "Engineering stability in gene networks by autoregulation", Nature 405: 590-593, 2000.
[10] Tuttle et al. "Model-Driven Designs of an Oscillating Gene Network", Biophys J 89(6):3873-3883, 2005.
[11] McMillen LM et al. "Synchronizing genetic relaxation oscillators by intercell signaling", P Natl Acad Sci USA 99(2):679-684, 2002.
[12] Basu S et al. "A synthetic multicellular system for programmed pattern formation", Nature 434:1130-1134, 2005.
[13] Einstein, Albert , "On the Motion—Required by the Molecular Kinetic Theory of Heat—of Small Particles Suspended in a Stationary Liquid", 1905.
[14] Einstein, Albert , "A new determination of molecular dimensions", 1905.
[15] Gierer A and Meinhardt H, "A theory of biologycal pattern formation", Kybernetik 12:30-39, 1972
[16] Turing AM, "The Chemical Basis of Morphogenesis", Philosophical Transactions of The Royal Society of London, series B, 237:37–72, 1952.
[17] Wolpert L, "Positional information and the spatial pattern of cellular differentiation", Journal of Theoretical Biology, 25:1–47, 1969
[18] Murray JD, "Mathematical Biology II", Springer-Verlag Berlin Heidelberg, 3rd edition, 1993





 "
"