Team:IPN-UNAM-Mexico/Project
From 2009.igem.org
m (→50pxReaction-Diffusion systems implemented on Biobricks) |
m (→50pxReaction-Diffusion systems implemented on Biobricks) |
||
| Line 132: | Line 132: | ||
| - | For a complete | + | For a complete documentation of the biobricks check [[Team:IPN-UNAM-Mexico/Parts | parts section]]. |
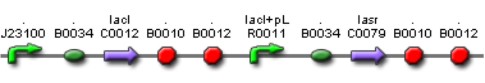
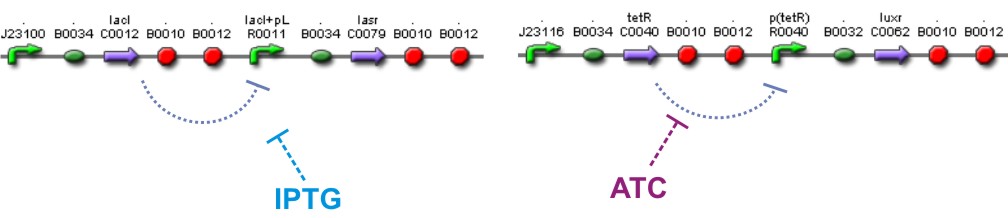
'''[[Team:IPN-UNAM-Mexico/Parts | Lac inverter]]''' | '''[[Team:IPN-UNAM-Mexico/Parts | Lac inverter]]''' | ||
[[Image:AI Lac.jpg|500px|center|Lac Inverter]] | [[Image:AI Lac.jpg|500px|center|Lac Inverter]] | ||
| + | |||
| + | |||
| + | The ''Lac ''Inverter is constitutive producing ''LacI ''and repressing ''plac ''promoter. ''Plac ''promoter controls de expression of ''LasR''. | ||
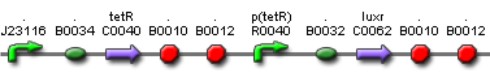
'''[[Team:IPN-UNAM-Mexico/Parts | Tet inverter]]''' | '''[[Team:IPN-UNAM-Mexico/Parts | Tet inverter]]''' | ||
[[Image:AI Tet.jpg|500px|center|link=https://2009.igem.org/Team:IPN-UNAM-Mexico/Parts]] | [[Image:AI Tet.jpg|500px|center|link=https://2009.igem.org/Team:IPN-UNAM-Mexico/Parts]] | ||
| + | |||
| + | |||
| + | The Tet'' ''Inverter is constitutive producing ''TetR ''and repressing ''ptet ''promoter. ''Ptet ''promoter controls de expression of ''LuxR''. | ||
| Line 146: | Line 152: | ||
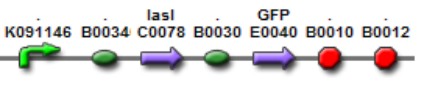
[[Image:AI LuxI.jpg|300px|center|Lux AHL]] | [[Image:AI LuxI.jpg|300px|center|Lux AHL]] | ||
| + | |||
| + | |||
| + | This biobrick is a signaling protein generator that is expected to be activated by ''LasR''+''PAI'' controlling the expression of ''LuxI'' enzyme. | ||
| + | |||
| + | |||
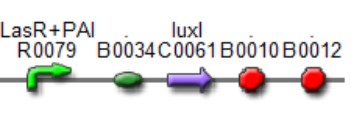
'''[[Team:IPN-UNAM-Mexico/Parts | Las AHL]]''' | '''[[Team:IPN-UNAM-Mexico/Parts | Las AHL]]''' | ||
| Line 152: | Line 163: | ||
| + | This biobrick is a signaling proteing generator that is expected to controlled in two different ways, activated by ''LasR''+''PAI'' and repressed by ''LuxR''+''AI.'' It controls the expression of LasI enzyme and GFP in a polycistronic way. | ||
Revision as of 20:37, 21 October 2009
Turing meets synthetic biology
 Introduction
Introduction
The work of Turing
In 1952 Alan M. Turing in his classical paper called The chemical basis of morphogenesis he sugested that "...a system of chemical substances, called morphogens, reacting together and diffusing through a tissue, is adequate to account for the main phenomena of morphogenesis." This system is amazing by its simplicity. With only two morphogens it's possible to reproduce nontrivial patterns that are similar to those of zebras or leopards.
Although there is not enough evidence of the existence of these morphogens in living organisms, the likeliness of the patterns obtained by theoretical means using this model with the ones found in nature is astonishing.
Turing assumed that the morphogenes can react with each other and diffuse through cells. It is necessary that at the beginning there is a non-homogeneous distribuition of these morphogenes, which is often called chemical prepattern and can be given merely by random disturbances. An intuitive notion would tell us that the diffusion of the morphogenes starting from this chemical prepattern would led to a homogenous state of the system, but surprisingly the Turing proposal says that non-homgenous structures will arise, as a direct consequence of diffusion (the turing hyphotesis); reaching a stable state with regions with high concentrantions of one morphogen and regions with high concentration of the others. This non-homogeneous distribuition patterns of morphogens resembles those found in nature during some stages of morhpogenesis (from the gastrulation or the tentacle patterns on hydras to the jaguar spots).
Reaction-Diffusion equations
Turing formally described his proposal with a set of Partial Differential Equations where is possible to represent the chemical interactions of the morphogens and the way they move over the space. This kind of dynamics are commonly called reaction-diffusion mechanisms, and the equations that describe them are named Reaction-Diffusion equations. In section modeling we formally present them.
The Activator-Inhibitor
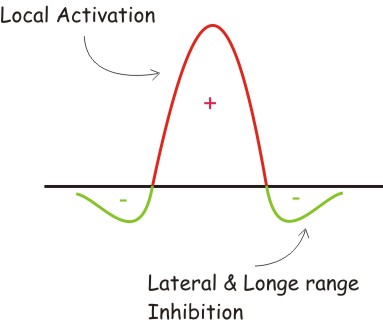
In 1972 A. Gierer and H. Meinhardt published his work called A theory of biological pattern formation presenting a network based in the interaction of at least two morphogenes acting as an activator and an inhibitor. The main qualitative dynamics of the morphogens is:
Short range activation, longer range inhibition and a conceptual distinction between effective concentrations of activator and inhibitor, on one hand, and the density of their sources on the other.
Schematically we can see this description as:
IMAGEN
J.D. Murray presented a good example of how patterns can arise from this kind of dynamics so we cite his text literally:
Synthesizing, we can now represent the interactions between the activator and the inhibitor as follows:
The activator is autocathalyzed and it also triggers the formation of the inhibitor. Accordingly to its name, the inhibitor inhibits the production of the activator, leading to a simple but very rich dymamics (For more details on the acivator-inhibitor equations please go to modelling section). From the grass field and the fire analogy we can deduce that the diffusion coefficients of the fire should be slower than the grasshoppers one; if not the fire (activator) would spread completely in the area. This is an important fact that we mention again later.
GENERAL CONDITIONS FOR PATTERN GENERATION
Although this kind of conditions refers mainly to a mathematical analyses of the reaction-diffusion systems like the Activator-Inhibitor of J.D. Murray , we can qualitative simplify and establish according to this analyses the general conditions for pattern formation:
- The existence of at least of two morphogenes with different nature that interacts chemically between them diffuse over the space
- The coefficient rates of diffusion should be different.
- The starting distribution of morphogenes should not be completely homogeneous over the space.
- The Gierer and Mainhardt proposal: local activation and longe range inhibition.
 TURING MEETS SYNTHETIC BIOLOGY
TURING MEETS SYNTHETIC BIOLOGY
The Synthetic Biology has become in the last years an excellent tool to designe biological systems with particular purposes. In our team we decided to used it as a way to contribute not only with technological aspects but mainly with theoretical and foundational research.
The Turing's ideas about cellular differentiation in zygote according to gradients of morphogenes that react and diffuse through the cellular tissue have not been completely confirmed due to the lack of biological evidence. The main objective of this project is to give an example of a biological system that contains reaction-diffusion mechanisms and is able to reproduce spatiotemporal Turing-type patterns or another kind of nontrivial patterns.
The biobricks found in the [http://www.partsregistry.org Registry of Biological parts] are a good starting point to construct a synthetic network of genes that due to their interactions could be identified as an Activator-Inhibitor system. This genetic network inserted innto an E. coli' should respond to the concentration gradient of morphogens in the media and have the potential to differentiate accordingly.
The main challenge was to design a synthetic network that fulfill the general conditions for generating a spatiotemporal pattern (see above). But we must act carefully, since in Murray's book says literaly:
an advise that few teams in the past years' competitions may have considered considered.
 Morphogenesis qualitative mechanisms
Morphogenesis qualitative mechanisms
 Reaction-Diffusion systems implemented on Biobricks
Reaction-Diffusion systems implemented on Biobricks
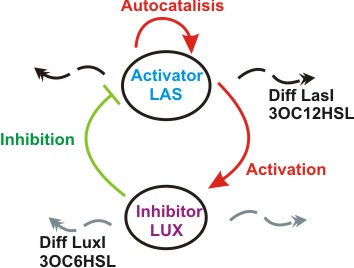
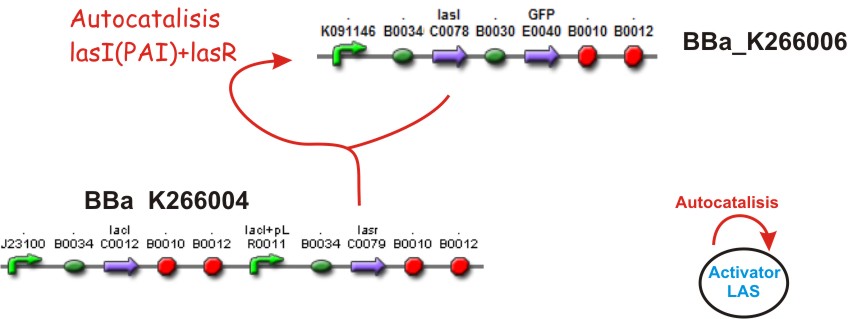
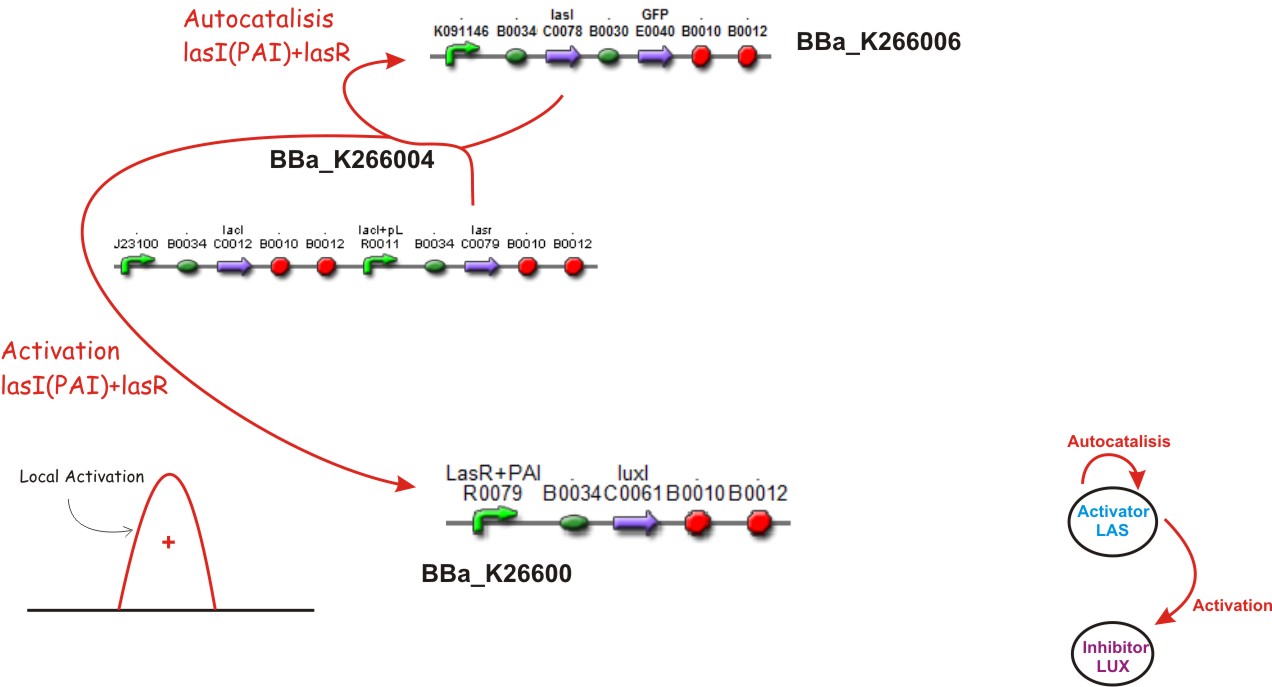
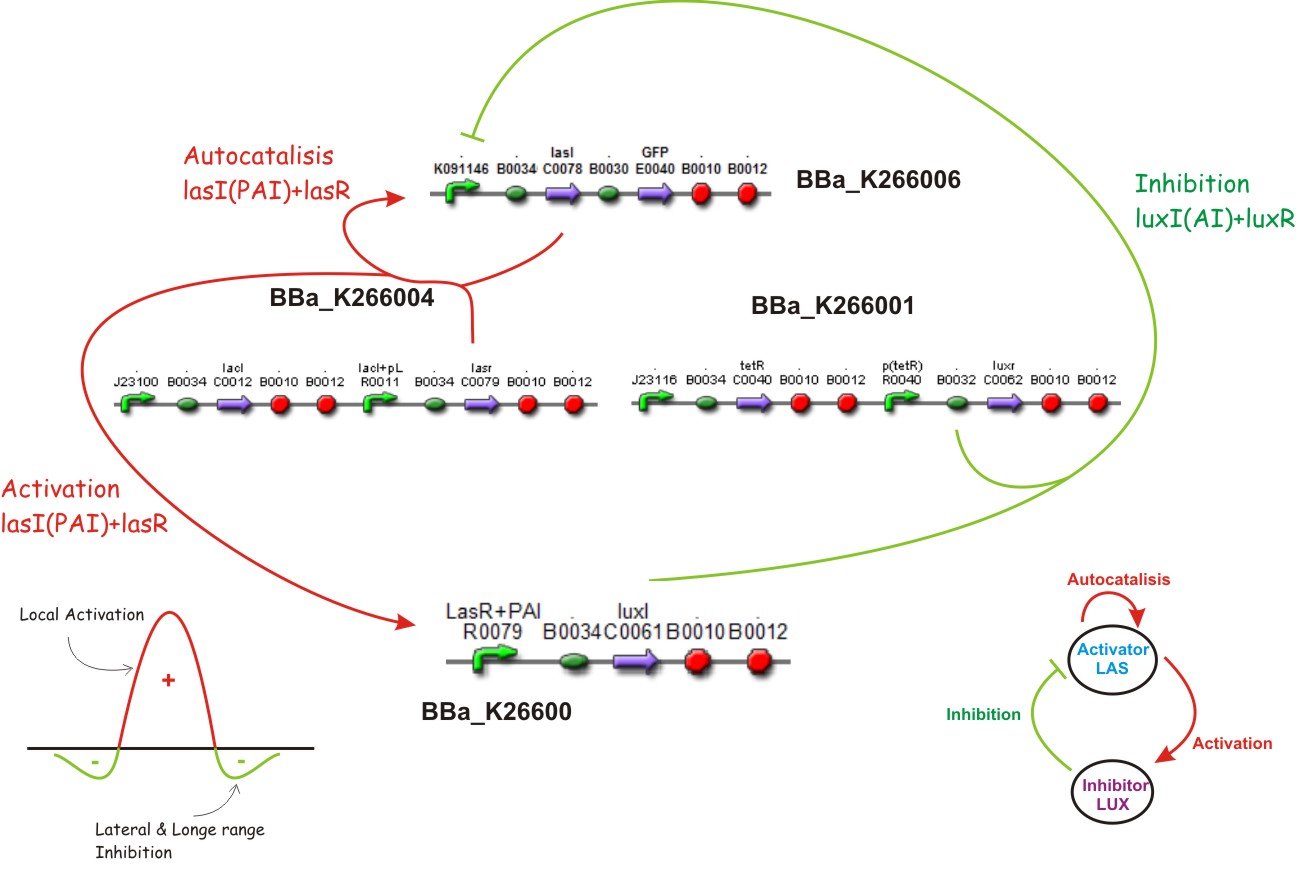
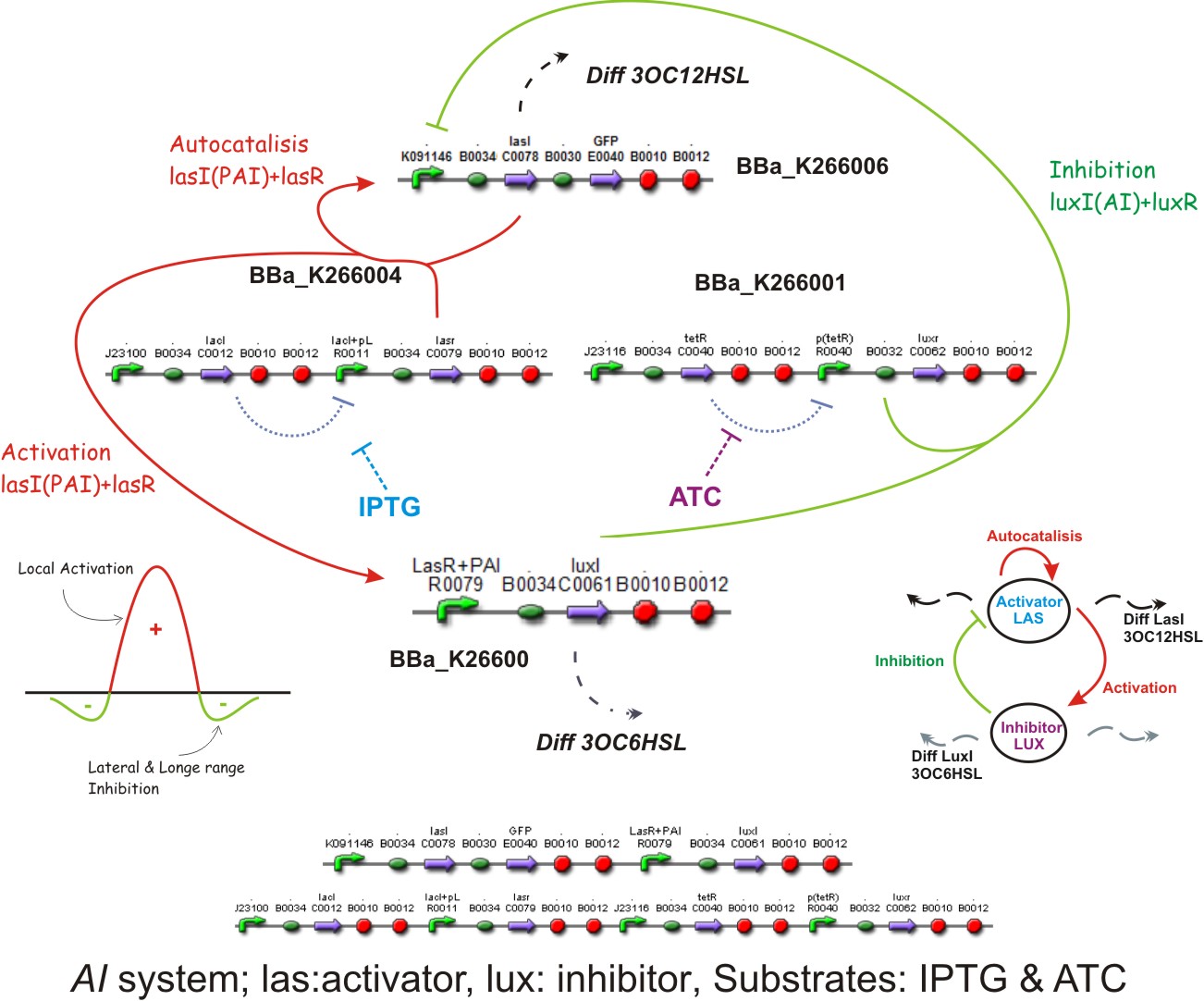
Inspired by the Gierer and Mainhardt work we designed a synthetic genetic network that acts as an Activator-Inhibitor with limiting substrates. We explain each particular characteristics of reaction, diffusion and substrates in the following subsections.
Reaction - Diffusion
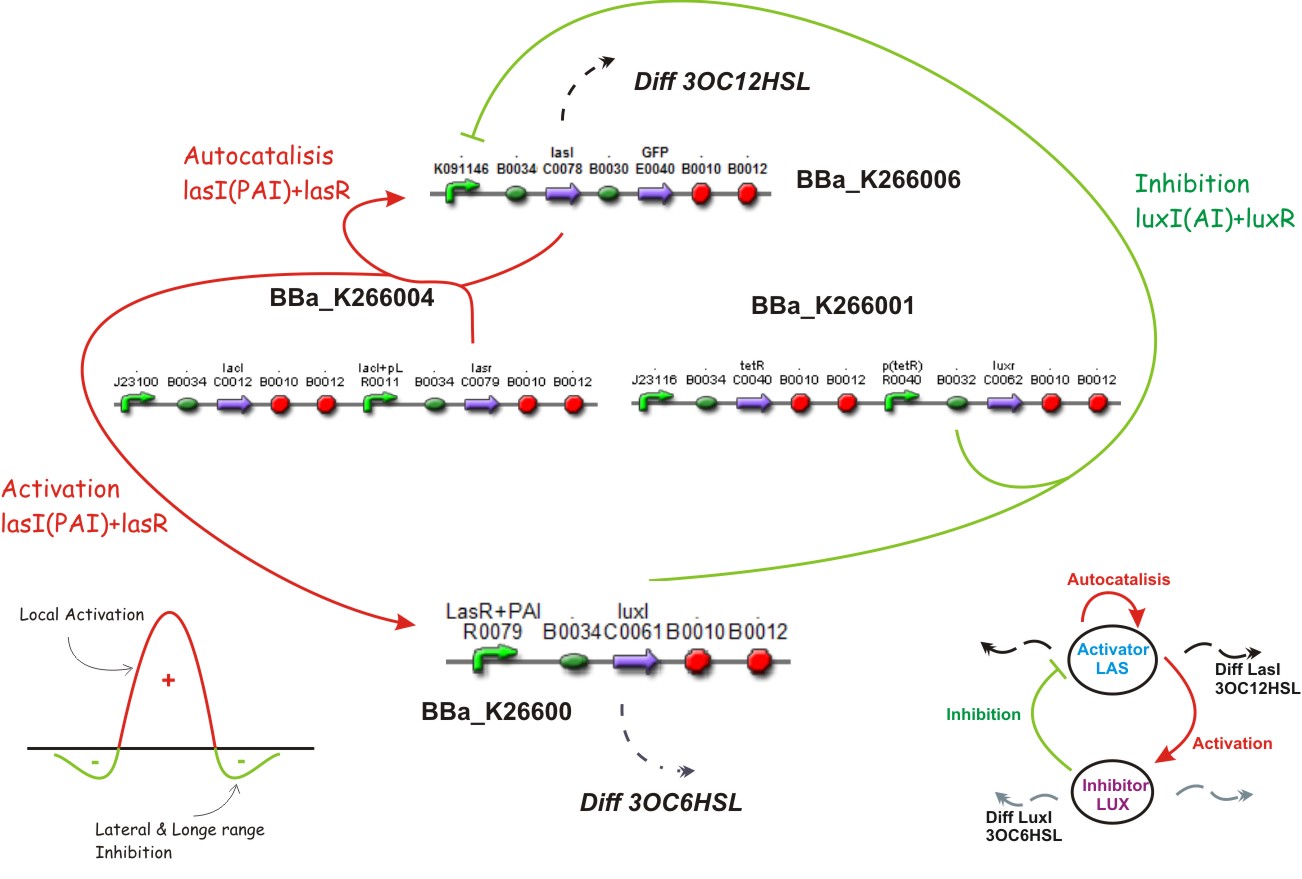
We suppose the existence of two morphogenes, represented by Lux and Las , AI and PAI lactones respectively. This substances are very small and travel throw the membrane, and diffuse fast in the media.
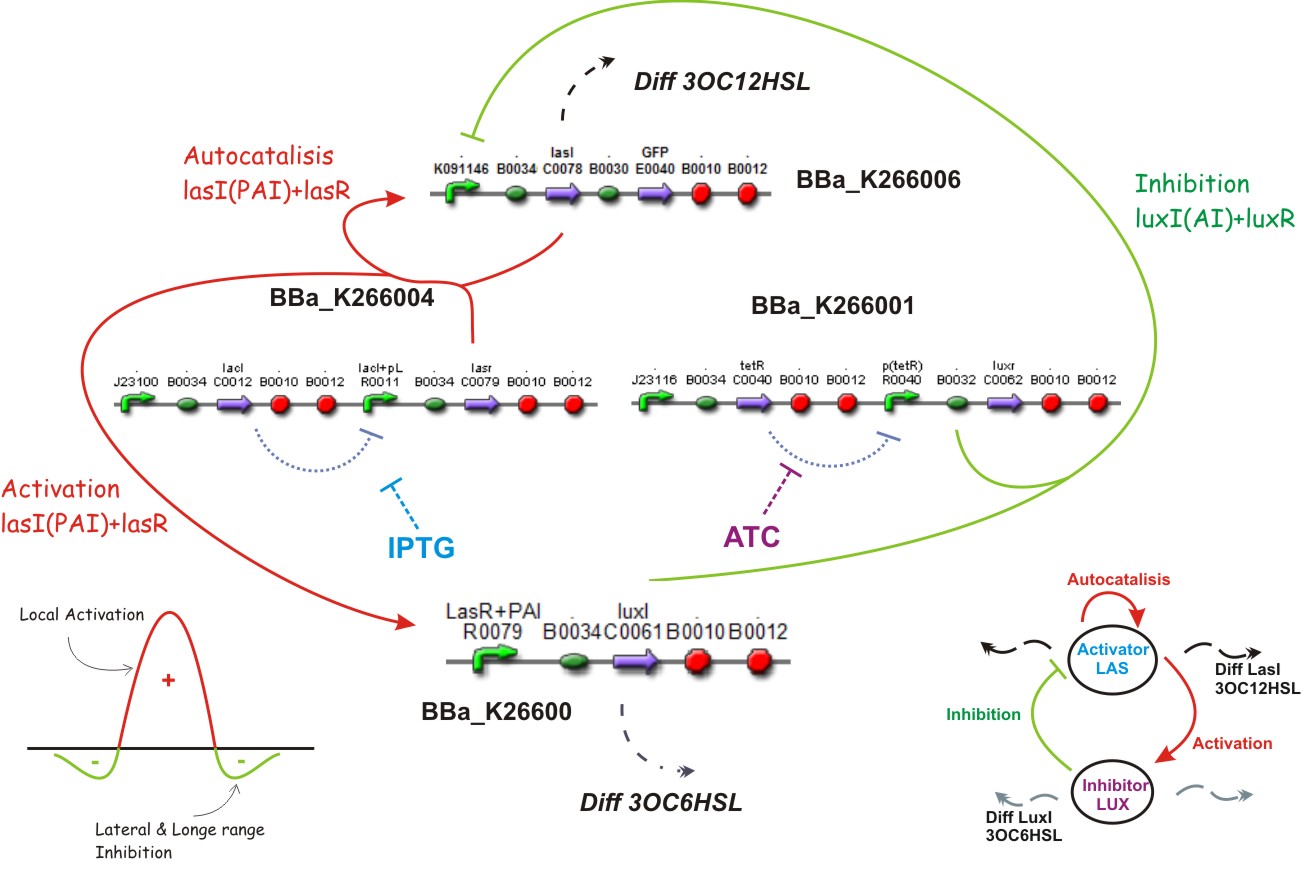
We used two promoters, one inducible by LasR+PAI ([http://www.partsregistry.org/Part:Bba_R0079 Bba_R0079]) and the other double controlled ([http://www.partsregistry.org/Part:Bba_RK091146 Bba_K091146]), repressed by LuxR+AI and induced by LasR+PAI; in our genetic network we gave the cell the posibility of responde to a gradient of PAI and AI and express GFP.
We used too a constitutive Lac Inverter, to control the production of LasR with IPTG, a constitutive Tet inverter that controls LuxR with ATC. These inverters provides a good way to control the systems and make it more sensitive for parameters adjust.
For a complete documentation of the biobricks check parts section.
The Lac Inverter is constitutive producing LacI and repressing plac promoter. Plac promoter controls de expression of LasR.
The Tet Inverter is constitutive producing TetR and repressing ptet promoter. Ptet promoter controls de expression of LuxR.
This biobrick is a signaling protein generator that is expected to be activated by LasR+PAI controlling the expression of LuxI enzyme.
This biobrick is a signaling proteing generator that is expected to controlled in two different ways, activated by LasR+PAI and repressed by LuxR+AI. It controls the expression of LasI enzyme and GFP in a polycistronic way.
Substrates
[http://openwetware.org/wiki/IPTG IPTG] & [http://openwetware.org/wiki/ATc ATC]
 The Activator-Inhibitor system on biobricks: The dynamics
The Activator-Inhibitor system on biobricks: The dynamics
Autocatalisis
Activation
Inhibition
Diffusion
Substrates
Complete system





 "
"