Team:Illinois/SgrS
From 2009.igem.org
(→SgrS Target-GFP Fusion) |
|||
| Line 9: | Line 9: | ||
'''Protocol(s) Used:''' (links to protocols page) | '''Protocol(s) Used:''' (links to protocols page) | ||
| + | |||
| + | Colony Boil-Streak plate E. Coli K12 on LB agar, incubate overnight at 37C. Pick healthy colony with sterile glass pasteur pipette and innoculate 100 uL dH20. Boil for 10 min, centrifugate for 5 min at 14K rpm. Extract 1 uL of supernatant as template DNA for PCR. | ||
'''Recipe(s) Used:''' | '''Recipe(s) Used:''' | ||
Revision as of 22:20, 18 June 2009
Contents |
SgrS Target-GFP Fusion
People in Group: Francis Lee, Harsh Shah, Dave Korenchan
Purpose: We want to fuse the SgrS target sequence from the PtsG gene to the GFP gene on a low-copy plasmid and transform this into E. coli cells along with a high-copy plasmid carrying the SgrS gene. We expect to see translational repression of GFP by the small RNA SgrS.
Protocol(s) Used: (links to protocols page)
Colony Boil-Streak plate E. Coli K12 on LB agar, incubate overnight at 37C. Pick healthy colony with sterile glass pasteur pipette and innoculate 100 uL dH20. Boil for 10 min, centrifugate for 5 min at 14K rpm. Extract 1 uL of supernatant as template DNA for PCR.
Recipe(s) Used:
sgrS Gene Sequence: (K-12 substr. MG1655: 77366-77623)
GATGA AGCAA GGGGG TGCCC CATGC GTCAG TTTTA TCAGC ACTAT TTTAC CGCGA CAGCG AAGTT GTGCT GGTTG CGTTG GTTAA GCGTC CCACA ACGAT TAACC ATGCT TGAAG GACTG ATGCA GTGGG ATGAC CGCAA TTCTG AAAGT TGACT TGCCT GCATC ATGTG TGACT GAGTA TTGGT GTAAA ATCAC CCGCC AGCAG ATTAT ACCTG CTGGT TTTTT TTATT CTCGC CGCGC TAAAA AGGGA ACGTA TG
ptsG 5' UTR Sequence: (K-12 substr. MG1655: 1156990-1157166)
AAATA AAGGG CGCTT AGATG CCCTG TACAC GGCGA GGCTC TCCCC CCTTG CCACG CGTGA GAACG TAAAA AAAGC ACCCA TACTC AGGAG CACTC TCAAT TATGT TTAAG AATGC ATTTG CTAAC CTGCA AAAGG TCGGT AAATC GCTGA TGCTG CCGGT ATCCG TACTG CCTAT C
Primers Used:
- Forward/sense sgrS (sRNA gene) primer: 5'p+GATGAAGCAAGGGGGTGCCC
- Reverse/antisense sgrS (sRNA gene) primer: GTTTTTTCTAGACATACGTTCCCTTTTTAGCG
- Forward/sense ptsG (sRNA target sequence) primer: GTTTTTATGCATAAATAAAGGGCGCTTAGATG
- Reverse/sense ptsG (sRNA target sequence) primer: GTTTTTGCTAGCGATAGGCAGTACGGATAC
PCR Products:
sgrS GENE (XbaI restriction site)-269bp:
GATGA AGCAA GGGGG TGCCC CATGC GTCAG TTTTA TCAGC ACTAT TTTAC CGCGA CAGCG AAGTT GTGCT GGTTG CGTTG GTTAA GCGTC CCACA ACGAT TAACC ATGCT TGAAG GACTG ATGCA GTGGG ATGAC CGCAA TTCTG AAAGT TGACT TGCCT GCATC ATGTG TGACT GAGTA TTGGT GTAAA ATCAC CCGCC AGCAG ATTAT ACCTG CTGGT TTTTT TTATT CTCGC CGCGC TAAAA AGGGA ACGTA TGTCT AGAAA AAAC
ptsG 5' UTR (Mph1103I, NheI restriction sites)-200bp:
GTTTT TATGC ATAAA TAAAG GGCGC TTAGA TGCCC TGTAC ACGGC GAGGC TCTCC CCCCT TGCCA CGCGT GAGAA CGTAA AAAAA GCACC CATAC TCAGG AGCAC TCTCA ATTAT GTTTA AGAAT GCATT TGCTA ACCTG CAAAA GGTCG GTAAA TCGCT GATGC TGCCG GTATC CGTAC TGCCT ATCGC TAGCA AAAAC
June 15
sgrS Gene and ptsG 5'UTR were PCR-cloned out of the chromosome using colony boil. sgrS Gene was cloned with 3' XbaI restriction site. ptsG Gene was cloned with 5' Mph1103I restriction site and a 3' NheI restriction site.
June 16
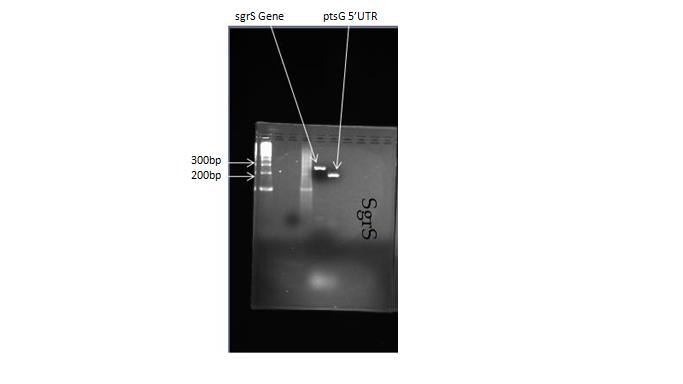
We ran a 3% agarose gel on our PCR reactions, and the PCRs were successful! The SgrS gene falls just under the 300 bp region on our gel, which agrees with our data for the length of the SgrS gene segment. The PtsG target sequence band is located at 200bp, which also agrees with our data for the length of the ptsG target sequence with its respective fused restriction sites. Reaction was purified with Nucleospin Extract DNA kit and stored at -20C.
June 17
Both sgrS Gene and ptsG 5'UTR were digested with restriction enzymes- XbaI and Mph1103I/NheI respectively. Reaction was resolved on 2% agarose gel, which was subsequently extracted and purified with the Nucleospin Extract DNA kit. Final elution (15uL) was stored at -20C
 "
"