Team:Illinois/Vectors
From 2009.igem.org
(→Gels) |
(→Gels) |
||
| Line 9: | Line 9: | ||
This is the second gel of the PCR that we did of the pJU-334 DNA sequence. The band shows up at ~11 kbp --oops. Again, we expected a band at ~3.1 kbp. | This is the second gel of the PCR that we did of the pJU-334 DNA sequence. The band shows up at ~11 kbp --oops. Again, we expected a band at ~3.1 kbp. | ||
| - | [[Image: | + | [[Image:Illinois-_Gel_2_of_PCR_of_pJU-334.jpg]] |
This is the third gel of the PCR that we did of the pJU-334 DNA sequence. Here we see two band. A faint one, (where we expected the band to show up) at 3.1 kbp. The second, more prominent band is at ~1.9 kpb again. | This is the third gel of the PCR that we did of the pJU-334 DNA sequence. Here we see two band. A faint one, (where we expected the band to show up) at 3.1 kbp. The second, more prominent band is at ~1.9 kpb again. | ||
| - | [[Image: | + | [[Image:Illinois-_Gel_3_of_pJU-334.jpg]] |
This is the fourth gel of the PCR that we did of the pJU-334 DNA sequence. This time, we put in all 45 micro liters of the DNA. Again, we got fainter bands at 3.1 kbp and stronger ones at ~1.9. Then we purified the 3.1 kbp fragments of DNA. We then used this purified DNA for another PCR. Next, we intend to run a gel of this, isolate the 3.1 kbp fragment, and use this to proceed with our experiment. | This is the fourth gel of the PCR that we did of the pJU-334 DNA sequence. This time, we put in all 45 micro liters of the DNA. Again, we got fainter bands at 3.1 kbp and stronger ones at ~1.9. Then we purified the 3.1 kbp fragments of DNA. We then used this purified DNA for another PCR. Next, we intend to run a gel of this, isolate the 3.1 kbp fragment, and use this to proceed with our experiment. | ||
| - | [[Image: | + | [[Image:Illinois-_Gel_4_of_pJU-334.jpg]] |
Revision as of 19:11, 19 June 2009
Contents |
Gels
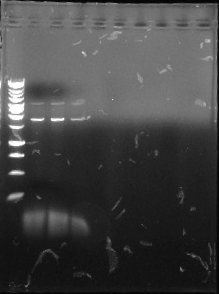
This is the first gel of the PCR that we did of the pJU-334 DNA sequence. The band shows up at ~1.9 kbp. We expected it to be at 3.1 kbp.
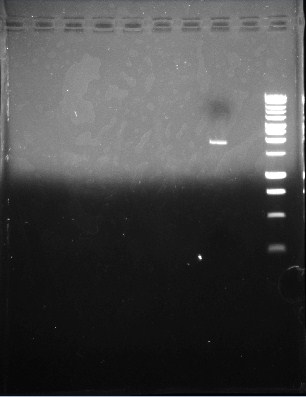
This is the second gel of the PCR that we did of the pJU-334 DNA sequence. The band shows up at ~11 kbp --oops. Again, we expected a band at ~3.1 kbp.
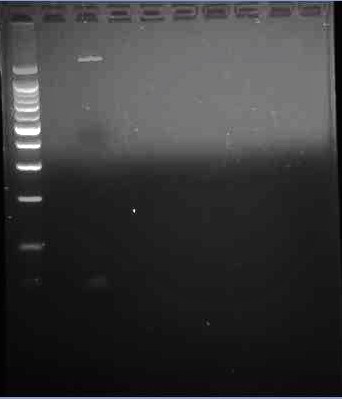
This is the third gel of the PCR that we did of the pJU-334 DNA sequence. Here we see two band. A faint one, (where we expected the band to show up) at 3.1 kbp. The second, more prominent band is at ~1.9 kpb again.
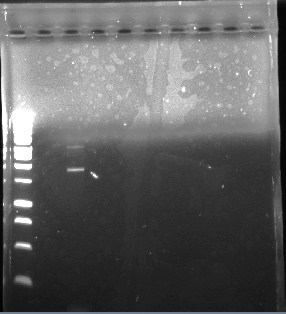
This is the fourth gel of the PCR that we did of the pJU-334 DNA sequence. This time, we put in all 45 micro liters of the DNA. Again, we got fainter bands at 3.1 kbp and stronger ones at ~1.9. Then we purified the 3.1 kbp fragments of DNA. We then used this purified DNA for another PCR. Next, we intend to run a gel of this, isolate the 3.1 kbp fragment, and use this to proceed with our experiment.
Update: we just decided that maybe they told us to use the worng primer. So the question becomes, how come we got DNA fragments that were about the right size? Maybe the primers similar.
Vectors
This page contains maps of the plasmids used in our project.
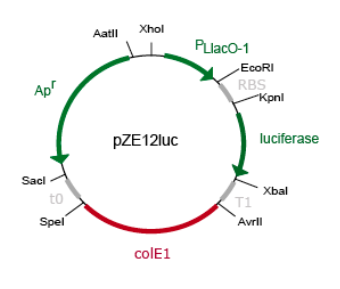
PJU-334 Plasmid
Note: The PZE12luc plasmid is the parental plasmid of the PJU-334 linear fragment.
 "
"