Team:Kyoto/GSDD/Parts
From 2009.igem.org
(Difference between revisions)
ShoTakamori (Talk | contribs) m |
|||
| (6 intermediate revisions not shown) | |||
| Line 21: | Line 21: | ||
#[[Team:Kyoto/GSDD|GSDD]] | #[[Team:Kyoto/GSDD|GSDD]] | ||
#Parts | #Parts | ||
| + | {{:Team:Kyoto/topicpath}} | ||
</div><!-- /#topicpath --> | </div><!-- /#topicpath --> | ||
{{:Team:Kyoto/Leftnavi}} | {{:Team:Kyoto/Leftnavi}} | ||
| Line 30: | Line 31: | ||
We designed following three vectors and registered intermediate products of Timer vector and constitutive LacI generator optimized in yeast. | We designed following three vectors and registered intermediate products of Timer vector and constitutive LacI generator optimized in yeast. | ||
| - | ==Timer Vector for functional measurement experiment== | + | ===Timer Vector for functional measurement experiment=== |
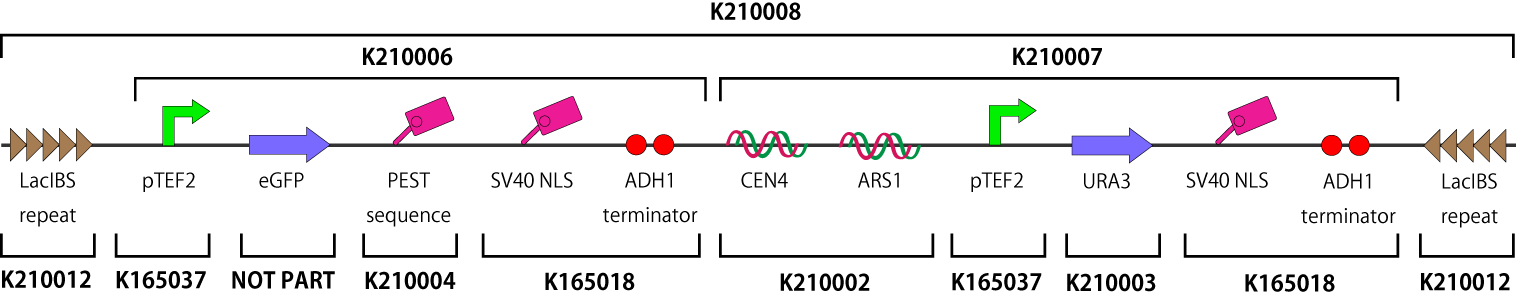
| - | This part is designed for our functional measurement of Timer vector. It consists of four part (see figure); both ends of repeated sequence of LacI binding site, constitutive eGFP generator (pTEF2-kozac-eGFP) with degradation promoted sequence (PEST), yeast centromeric sequence (CEN), yeast replication origin (ARS), constitutive uracil generator (pTEF2-kozac-URA3) with SV40 nuclear localization sequences and two ADH1 terminators (SV40-ADH1). As mentioned in Mechanism, Timer vector is a linear DNA and inserted into pSB1A2. When we use this, we can cut out linear Timer vector with a pair of restriction enzyme frequently used in iGEM, (EcoRI or XbaI)and(SpeI or PstI).[[Image:figure(timer).png|740px|center|Fig.1]] | + | This part is designed for our functional measurement of Timer vector. It consists of four part (see figure); both ends of repeated sequence of LacI binding site, constitutive eGFP generator (pTEF2-kozac-eGFP) with degradation promoted sequence (PEST), yeast centromeric sequence (CEN), yeast replication origin (ARS), constitutive uracil generator (pTEF2-kozac-URA3) with SV40 nuclear localization sequences and two ADH1 terminators (SV40-ADH1). As mentioned in Mechanism, Timer vector is a linear DNA and inserted into pSB1A2. When we use this, we can cut out linear Timer vector with a pair of restriction enzyme frequently used in iGEM, (EcoRI or XbaI)and(SpeI or PstI).[[Image:figure(timer)2.png|740px|center|Fig.1]] |
| - | ==Constitutive LacI generator== | + | ===Constitutive LacI generator=== |
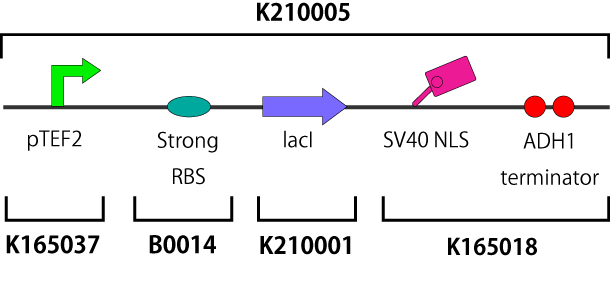
| - | This part is also designed for our experiment. It consists of constitutive LacI generator (pTEF2-kozac-LacI) with SV40 nuclear localization sequences and two ADH1 terminators (SV40-ADH1). This part is also inserted into | + | This part is also designed for our experiment. It consists of constitutive LacI generator (pTEF2-kozac-LacI) with SV40 nuclear localization sequences and two ADH1 terminators (SV40-ADH1). This part is also inserted into BYP5005(national bioresource project).[[Image:Figure(lacI_generator)2.png|300px|center|Fig.1]] |
</div><!-- /#rightcontents --> | </div><!-- /#rightcontents --> | ||
Latest revision as of 02:55, 22 October 2009
Parts
Parts Design
We designed following three vectors and registered intermediate products of Timer vector and constitutive LacI generator optimized in yeast.
 "
"