Team:Minnesota/Parts Characterization
From 2009.igem.org
| Line 1: | Line 1: | ||
| - | {|align="center" cellpadding="25" | + | {|align="center" cellpadding="25" |[[Image:mnlogo.jpg|300px|center|frame]] |} {| style="color:gold;background-color:#800000;" cellpadding="3" cellspacing="1" border="1" bordercolor="#fff" width="90%" align="center" !align="center"|[[Team:Minnesota|<font color="gold">Home</font>]] !align="center"|[[Team:Minnesota/Team|<font color="gold">The Team</font>]] !align="center"|[[Team:Minnesota/Project|<font color="gold">The Project</font>]] !align="center"|[[Team:Minnesota/Parts|<font color="gold">Submitted Parts</font>]] !align="center"|[[Team:Minnesota/Modeling|<font color="gold">Modeling</font>]] !align="center"|[[Team:Minnesota/Designer|<font color="gold">SynBioSS Designer</font>]] !align="center"|[[Team:Minnesota/Parts Characterization|<font color="gold">Parts Characterization</font>]] !align="center"|[[Team:Minnesota/Notebook|<font color="gold">Experiments and Calendar</font>]] |} |
| - | |[[Image:mnlogo.jpg|300px|center|frame]] | + | <h1>Parts Characterization</h1> |
| - | |} | + | <h2>Introduction</h2> |
| + | After an exhaustive search of the [http://www.partsregistry.org/Main_Page Registry of Standard Parts], we found five promoters that were included in the 2009 iGEM Kit as potential parts characterize. Using part [http://partsregistry.org/wiki/index.php/Part:BBa_F2620 BBa_F2620], which was characterized by a group from MIT in 2004, as a template, we examined the following parts: | ||
| + | <table> | ||
| + | <table border="1"> | ||
| + | <tr> | ||
| + | <th>Part</th><th>Description</th><th>Regulators</th><th>People</th> </tr> <tr> <td>[http://partsregistry.org/Part:BBa_I14032 I14032]</td><td>Constitutive promoter classified as repressible</td><td>IPTG</td><td>Princeton 2004</td> </tr> <tr> <td>[http://partsregistry.org/Part:BBa_J13002 J13002]</td><td>Two TetR binding sites and RBS</td><td>aTc</td><td>UT Austin 2005</td> </tr> <tr> <td>[http://partsregistry.org/Part:BBa_I14015 I14015]</td><td>LasR, 3OC12HSL aTc regulated promoter</td><td>LasR, 3C12HSL, aTc</td><td>Princeton 2004</td> </tr> <tr> <td>[http://partsregistry.org/Part:BBa_K091101 K091101]</td><td>TTL AND gate</td><td>IPTG, aTc</td><td>Davidson Missouri-Western 2008</td> </tr> <tr> <td>[http://partsregistry.org/Part:BBa_R0011 R0011]</td><td>Inverting regulatory region controlled by LacI; for comparison since already characterized</td><td>IPTG</td><td>Registry</td> </tr> </table> | ||
| - | + | We chose these parts because they had the same regulators as the promoters we examined in our project. Part [http://partsregistry.org/Part:BBa_K091101 K091101] was particularly interesting because it was one of the constructs of the Tet and Lac operators that we examined for our research. Since Tet and Lac are commonly studied operators in synthetic biology, we wanted parts that involved them to be well-characterized to ensure the viability of future research. We decided to characterize these promoters by attaching them to part [http://partsregistry.org/Part:Bba_K081012 K081012], which consists of a strong RBS and GFP. This 'PoPS generator' takes [http://parts2.mit.edu/wiki/index.php/Abstraction_hierarchy_and_PoPS PoPS] (Polymerase Per Second) as an input and gives GFP as an output, allowing us to indirectly measure PoPS and characterize our parts. [[Image:BiobrickVector.jpg|500px|right]] The picture at the right, from [http://www.jbioleng.org/content/2/1/5 Shetty et al. Journal of Biological Engineering 2008] demonstrates how to combine standard biological parts to form a new composite part. In our case, the prefix part was each of our 5 promoters and these were digested with restriction enzymes EcoRI and SpeI. The suffix part was the PoPS generator in every case except part [http://partsregistry.org/Part:BBa_J13002 J13002], which already contained an RBS. This [http://partsregistry.org/wiki/index.php?title=Part:BBa_B0034 RBS] is defined as efficiency 1.0 while the RBS contained on our PoPS generator, which has an efficiency of 0.6. We decided to characterize the entire part that [http://parts2.mit.edu/wiki/index.php/UT_Austin_2005 UT Austin 2005] submitted, which included their RBS. These suffix parts were cut using restriction enzymes XbaI and PstI. We ligated these parts to make a composite BioBrick part in [http://partsregistry.org/Part:pSB3K3 pSB3K3], a low-medium copy plasmid with kanamycin resistance. The [https://2009.igem.org/Judging/Judging_Criteria 2009 iGEM Judging Criteria] gives MIT's characterization of part [http://partsregistry.org/wiki/index.php/Part:BBa_F2620 BBa_F2620] as an exemplar of parts characterization. We gratefully acknowledge their pioneering work with parts characterization and hope that our work continues to maintain the high standard for characterization of parts. We characterized our 5 promoters included with the 2009 iGEM kit in terms of: <ol> <li>Transfer Function: the equilibrium relationship between the input and output</li> <li>Specificity: the ability of the devide to distinguish between its true input and similar inputs</li> <li>Response time: the time taken for the output to respond to a change in input</li> <li>Stability: how transfer function changes across multiple rounds of cell division and culture</li> </ol> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <h2>Methods in the Lab</h2> | |
| + | <h4>Making the New Parts</h4> | ||
| + | Initially, we resuspended the DNA for the promoters, PoPS generator and GFP in water and [https://2009.igem.org/Team:Minnesota/Notebook transformed] them into [http://openwetware.org/wiki/TOP10_chemically_competent_cells Top10 chemically competent cells]. The plasmid backbone we transformed into CCDB-resistant cells. Then, we allowed the cells to grow overnight on the appropriate antibiotic plate based on the plasmid that the part was on. We picked colonies and inoculated liquid media. Once these cultures entered [http://en.wikipedia.org/wiki/Stationary_phase_(biology) stationary phase], we prepped the plasmids using the [https://2009.igem.org/Team:Minnesota/Notebook QIAprep Spin Miniprep Kit] for each promoter, the PoPS generator, GFP and the plasmid backbone (psB3K3) into which everything would be ligated. We quantified the purity of our DNA before performing polymerase chain reaction [https://2009.igem.org/Team:Minnesota/Notebook (PCR)] to amplify the DNA we had from the plasmid prep. Once the PCR completed, we usually ran some of the products out on an agarose gel to ensure that our DNA was the right size. We also sent some of our DNA to be sequenced and gratefully acknowledge the [http://www.bmgc.umn.edu/facilities/home.html BioMedical Genomics Center (BMGC)] at the University of Minnesota for their resources and expertise. The gel and sequencing helped us ensure that we had the correct plasmids. We performed the restriction enzyme digest on PCR products and adjusted the reaction conditions based on the concentration of the DNA from the plasmid prep. This reaction ran for between 2 and 3 hours. We ran these products out on an agarose gel and excised the DNA with razor blades. Then, we purified the DNA using a [https://2009.igem.org/Team:Minnesota/Project#Protocols:_Standard_techniques_that_we_used_in_the_wet_lab QIAquick Gel Extraction Kit]. The purified DNA we [https://2009.igem.org/Team:Minnesota/Notebook ligated] overnight at 16C. One of the challenges of this step was determining the appropriate ratio of insert to backbone because we were performing double insert rather than a single insert. The ratio for a single insert is 3:1 of insert to backbone. We still wanted to maximize insertion efficiency but minimize cancatamerization of inserts, so we ligated at a 6:1 ratio of inserts to backbone. We [https://2009.igem.org/Team:Minnesota/Notebook transformed] the ligation products into Top10 cells after their overnight ligation and plated them on LB + kanamycin plates since our plasmid backbone contained kanamycin resistance. This selected for non-transformants. The CCDB toxin built into the plasmid backbone also selected against uncut plasmid backbone and the gel purification step also allowed us to select the correct DNA. We grew these plates overnight and grew cultures from colonies that grew. Then, we were able to characterize the parts in terms of the four areas above: transfer function, specificity, response time, and stability. | ||
| - | < | + | <br /> |
| - | We | + | <h4>Sequencing</h4> |
| - | + | We utilized the software [http://www.genecodes.com/ Sequencher] through the Minnesota Supercomputing Institute [https://www.msi.umn.edu/ (MSI)] to trim the ends of sequences we received from the BMGC and align them. A chromatogram, shown below, told us the strength of the signal for each base pair. A good sequencing reaction will have a single peak for each base pair while problems with the primer or multiple products in the sequencing reaction will result in multiple peaks with equally strong signals for each base pair. [[Image:Chromatogram.jpg|400px|left]] We wanted to compare both sequencing reactions we had as of 16 July 2009 to each other and to the sequences of the parts that were available on the [http://partsregistry.org/Main_Page Registry]. We obtained successful alignments for parts I14032, J13002 and K091101, which were regulated by IPTG, aTc and both inducers, respectively. It is interesting to note that while our samples appeared to have multiple products, that is, on the chromatogram that accompanied our sequence output, we saw multiple signals for each base pair, the sequence products not only aligned with each other, but with the sequences on the Registry. So we ordered new primers for another sequencing reaction and, based on these alignments as well as the fact that part R0011 had already been characterized and I14013 was regulated by LasR as well as aTc, we reduced our number of parts to characterize to three. <br /> | |
| - | + | <h2>Characterization Experiments</h2> | |
| - | + | Our list of parts to characterize was then narrowed down to: <ol> <li>[http://partsregistry.org/Part:BBa_I14032 I14032]</li> <li>[http://partsregistry.org/Part:BBa_J13002 J13002]</li> <li>[http://partsregistry.org/Part:BBa_K091101 K091101]</li> </ol> <br /> <h4>Promoter J13002: aTc regulation</h4> The first promoter we examined was [http://partsregistry.org/Part:BBa_J13002 J13002], which was submitted to the Registry by [http://parts.mit.edu/wiki/index.php/UT_Austin_2005 UT Austin] in 2005. The part consists of two TetR binding sites and RBS and is called a "TetR repressed PoPs/RIPS generator." Since the part already had a RBS, we attached [http://partsregistry.org/Part:BBa_E0040 GFP] in order to indirectly measure PoPS. A photo of this part from the [http://parts.mit.edu/ Registry of Standard Parts] is displayed below: [[Image:J13002.jpg|700px|left]] <br /> <br /> <br /> <br /> <br /> <br /> <br /> <br /> We inserted the promoter and GFP into the psB3K3 plasmid backbone in Top10 cells and confirmed our ligation by agarose gel and sequencing. Then, we transformed our new plasmid into DH5αPro cells, which constitutively express aTc and IPTG for characterization. Therefore, even in our inducer media of 0 ng/ml, we observed some fluorescence. The plasmid also conferred kanamycin resistance upon the cells so we plated them on LB + Kan media in order to select for our successful transformants. | |
| - | + | <br/ > | |
| - | + | We wanted to examine Transfer Function and Stability in this promoter. To do this, we grew 1 ml liquid cultures in different concentration of inducer media: 0, 1, 10, 50, 100 and 200 ng/ml of aTc. We sampled each of our cultures every hour for 9 hours, fixing the samples in 4% PFA and resuspending in PBS in preparation for [http://en.wikipedia.org/wiki/Flow_cytometry flow cytometry]. Flow cytometry allowed us to pick out the population of living cells and determine the amount of GFP that each cell was producing. | |
| - | + | [[Image:FACS.jpg|200px|left|A flow cytometer at UMN: http://www.immunology.umn.edu/immunology/prod/groups/med/@pub/@med/documents/asset/med_56297.jpg]] | |
| - | + | We utilized the software [http://www.flowjo.com/ FlowJo] to analyze our flow cytometry data. We were interested in the mean GFP for 100,000 cells at each timepoint and aTc concentration and were able to extract these data from the plethora of information included in the data file. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | The | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | </ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | We [ | + | |
<br /> | <br /> | ||
<h2>Challenges</h2> | <h2>Challenges</h2> | ||
| - | A day by day catalog of what we did for parts characterization can be found on our Google calendar on the [https://2009.igem.org/Team:Minnesota/Notebook Experiments and Calendar] page. Clearly, despite the summary above, some of these steps we had to redo over and over. Often, our DNA after plasmid preparation and restriction enzyme digest was not very pure and had a low concentration, which necessitated picking colonies, growing more cultures and ]prepping more plasmids. Sequencing, which was performed by the [http://www.agac.umn.edu/ BMGC] on the U of MN campus, provided us with a huge challenge because often, our sequences were not clean so we were not sure that we had the correct parts. | + | A day by day catalog of what we did for parts characterization can be found on our Google calendar on the [https://2009.igem.org/Team:Minnesota/Notebook Experiments and Calendar] page. Clearly, despite the summary above, some of these steps we had to redo over and over. Often, our DNA after plasmid preparation and restriction enzyme digest was not very pure and had a low concentration, which necessitated picking colonies, growing more cultures and ]prepping more plasmids. Sequencing, which was performed by the [http://www.agac.umn.edu/ BMGC] on the U of MN campus, provided us with a huge challenge because often, our sequences were not clean so we were not sure that we had the correct parts. [[Image:Ligation_Gel.jpg|400px|right|This is a gel of our successful ligations for Promoters 1, 2, and 5]] Running samples out on a gel as shown to the right, and specing samples can be very helpful in double-checking the size and purity of fragments, but sequencing was a very important step that really told us whether our procedure was viable. When we received wonky results-- that is, the signals were mixed and the samples appeared to have multiple products-- we hypothesized that this was due to the primer setting down in the wrong location during the sequence reaction or perhaps a hairpin loop structures getting in the way of the sequencing (we gratefully acknowledge John Barrett for his suggestions). <br /> |
| - | [[Image:Ligation_Gel.jpg|400px|right|This is a gel of our successful ligations for Promoters 1, 2, and 5]] | + | |
| - | Running samples out on a gel as shown to the right, and specing samples can be very helpful in double-checking the size and purity of fragments, but sequencing was a very important step that really told us whether our procedure was viable. When we received wonky results-- that is, the signals were mixed and the samples appeared to have multiple products-- we hypothesized that this was due to the primer setting down in the wrong location during the sequence reaction or perhaps a hairpin loop structures getting in the way of the sequencing (we gratefully acknowledge John Barrett for his suggestions). | + | |
| - | <br /> | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | <h2>Acknowledgements</h2> | ||
| + | To the MIT 2004 team for their pioneering work in Parts Characterization. Thanks also to the BMGC at UMN for sequencing our myriad of parts and the Masonic Cancer Center at UMN for allowing us to utilize the flow cytometers. We also acknowledge John Barrett for his help using FlowJo and suggestions on obtaining successful sequences. | ||
| - | <h2>References</h2> | + | <h2>References</h2> Shetty, R.P.; Endy, D.; Knight, T.F. 2008. Engineering BioBrick vectors from BioBrick parts. Journal of Biological Engineering 2. |
| - | Shetty, R.P.; Endy, D.; Knight, T.F. | + | |
Revision as of 20:09, 30 July 2009
Contents |
Parts Characterization
Introduction
After an exhaustive search of the [http://www.partsregistry.org/Main_Page Registry of Standard Parts], we found five promoters that were included in the 2009 iGEM Kit as potential parts characterize. Using part [http://partsregistry.org/wiki/index.php/Part:BBa_F2620 BBa_F2620], which was characterized by a group from MIT in 2004, as a template, we examined the following parts:
| Part | Description | Regulators | People |
|---|---|---|---|
| [http://partsregistry.org/Part:BBa_I14032 I14032] | Constitutive promoter classified as repressible | IPTG | Princeton 2004 |
| [http://partsregistry.org/Part:BBa_J13002 J13002] | Two TetR binding sites and RBS | aTc | UT Austin 2005 |
| [http://partsregistry.org/Part:BBa_I14015 I14015] | LasR, 3OC12HSL aTc regulated promoter | LasR, 3C12HSL, aTc | Princeton 2004 |
| [http://partsregistry.org/Part:BBa_K091101 K091101] | TTL AND gate | IPTG, aTc | Davidson Missouri-Western 2008 |
| [http://partsregistry.org/Part:BBa_R0011 R0011] | Inverting regulatory region controlled by LacI; for comparison since already characterized | IPTG | Registry |
- Transfer Function: the equilibrium relationship between the input and output
- Specificity: the ability of the devide to distinguish between its true input and similar inputs
- Response time: the time taken for the output to respond to a change in input
- Stability: how transfer function changes across multiple rounds of cell division and culture
Methods in the Lab
Making the New Parts
Initially, we resuspended the DNA for the promoters, PoPS generator and GFP in water and transformed them into [http://openwetware.org/wiki/TOP10_chemically_competent_cells Top10 chemically competent cells]. The plasmid backbone we transformed into CCDB-resistant cells. Then, we allowed the cells to grow overnight on the appropriate antibiotic plate based on the plasmid that the part was on. We picked colonies and inoculated liquid media. Once these cultures entered [http://en.wikipedia.org/wiki/Stationary_phase_(biology) stationary phase], we prepped the plasmids using the QIAprep Spin Miniprep Kit for each promoter, the PoPS generator, GFP and the plasmid backbone (psB3K3) into which everything would be ligated. We quantified the purity of our DNA before performing polymerase chain reaction (PCR) to amplify the DNA we had from the plasmid prep. Once the PCR completed, we usually ran some of the products out on an agarose gel to ensure that our DNA was the right size. We also sent some of our DNA to be sequenced and gratefully acknowledge the [http://www.bmgc.umn.edu/facilities/home.html BioMedical Genomics Center (BMGC)] at the University of Minnesota for their resources and expertise. The gel and sequencing helped us ensure that we had the correct plasmids. We performed the restriction enzyme digest on PCR products and adjusted the reaction conditions based on the concentration of the DNA from the plasmid prep. This reaction ran for between 2 and 3 hours. We ran these products out on an agarose gel and excised the DNA with razor blades. Then, we purified the DNA using a QIAquick Gel Extraction Kit. The purified DNA we ligated overnight at 16C. One of the challenges of this step was determining the appropriate ratio of insert to backbone because we were performing double insert rather than a single insert. The ratio for a single insert is 3:1 of insert to backbone. We still wanted to maximize insertion efficiency but minimize cancatamerization of inserts, so we ligated at a 6:1 ratio of inserts to backbone. We transformed the ligation products into Top10 cells after their overnight ligation and plated them on LB + kanamycin plates since our plasmid backbone contained kanamycin resistance. This selected for non-transformants. The CCDB toxin built into the plasmid backbone also selected against uncut plasmid backbone and the gel purification step also allowed us to select the correct DNA. We grew these plates overnight and grew cultures from colonies that grew. Then, we were able to characterize the parts in terms of the four areas above: transfer function, specificity, response time, and stability.
Sequencing
We utilized the software [http://www.genecodes.com/ Sequencher] through the Minnesota Supercomputing Institute (MSI) to trim the ends of sequences we received from the BMGC and align them. A chromatogram, shown below, told us the strength of the signal for each base pair. A good sequencing reaction will have a single peak for each base pair while problems with the primer or multiple products in the sequencing reaction will result in multiple peaks with equally strong signals for each base pair. We wanted to compare both sequencing reactions we had as of 16 July 2009 to each other and to the sequences of the parts that were available on the [http://partsregistry.org/Main_Page Registry]. We obtained successful alignments for parts I14032, J13002 and K091101, which were regulated by IPTG, aTc and both inducers, respectively. It is interesting to note that while our samples appeared to have multiple products, that is, on the chromatogram that accompanied our sequence output, we saw multiple signals for each base pair, the sequence products not only aligned with each other, but with the sequences on the Registry. So we ordered new primers for another sequencing reaction and, based on these alignments as well as the fact that part R0011 had already been characterized and I14013 was regulated by LasR as well as aTc, we reduced our number of parts to characterize to three.Characterization Experiments
Our list of parts to characterize was then narrowed down to:- [http://partsregistry.org/Part:BBa_I14032 I14032]
- [http://partsregistry.org/Part:BBa_J13002 J13002]
- [http://partsregistry.org/Part:BBa_K091101 K091101]
Promoter J13002: aTc regulation
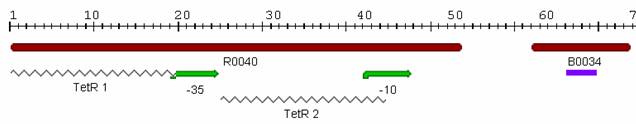
The first promoter we examined was [http://partsregistry.org/Part:BBa_J13002 J13002], which was submitted to the Registry by [http://parts.mit.edu/wiki/index.php/UT_Austin_2005 UT Austin] in 2005. The part consists of two TetR binding sites and RBS and is called a "TetR repressed PoPs/RIPS generator." Since the part already had a RBS, we attached [http://partsregistry.org/Part:BBa_E0040 GFP] in order to indirectly measure PoPS. A photo of this part from the [http://parts.mit.edu/ Registry of Standard Parts] is displayed below:We inserted the promoter and GFP into the psB3K3 plasmid backbone in Top10 cells and confirmed our ligation by agarose gel and sequencing. Then, we transformed our new plasmid into DH5αPro cells, which constitutively express aTc and IPTG for characterization. Therefore, even in our inducer media of 0 ng/ml, we observed some fluorescence. The plasmid also conferred kanamycin resistance upon the cells so we plated them on LB + Kan media in order to select for our successful transformants.
We wanted to examine Transfer Function and Stability in this promoter. To do this, we grew 1 ml liquid cultures in different concentration of inducer media: 0, 1, 10, 50, 100 and 200 ng/ml of aTc. We sampled each of our cultures every hour for 9 hours, fixing the samples in 4% PFA and resuspending in PBS in preparation for [http://en.wikipedia.org/wiki/Flow_cytometry flow cytometry]. Flow cytometry allowed us to pick out the population of living cells and determine the amount of GFP that each cell was producing.
We utilized the software [http://www.flowjo.com/ FlowJo] to analyze our flow cytometry data. We were interested in the mean GFP for 100,000 cells at each timepoint and aTc concentration and were able to extract these data from the plethora of information included in the data file.
Challenges
A day by day catalog of what we did for parts characterization can be found on our Google calendar on the Experiments and Calendar page. Clearly, despite the summary above, some of these steps we had to redo over and over. Often, our DNA after plasmid preparation and restriction enzyme digest was not very pure and had a low concentration, which necessitated picking colonies, growing more cultures and ]prepping more plasmids. Sequencing, which was performed by the [http://www.agac.umn.edu/ BMGC] on the U of MN campus, provided us with a huge challenge because often, our sequences were not clean so we were not sure that we had the correct parts. Running samples out on a gel as shown to the right, and specing samples can be very helpful in double-checking the size and purity of fragments, but sequencing was a very important step that really told us whether our procedure was viable. When we received wonky results-- that is, the signals were mixed and the samples appeared to have multiple products-- we hypothesized that this was due to the primer setting down in the wrong location during the sequence reaction or perhaps a hairpin loop structures getting in the way of the sequencing (we gratefully acknowledge John Barrett for his suggestions).Acknowledgements
To the MIT 2004 team for their pioneering work in Parts Characterization. Thanks also to the BMGC at UMN for sequencing our myriad of parts and the Masonic Cancer Center at UMN for allowing us to utilize the flow cytometers. We also acknowledge John Barrett for his help using FlowJo and suggestions on obtaining successful sequences.
 "
"