Team:Newcastle/Project/Labwork/Week1
From 2009.igem.org
Contents
|
Introductory Lab Sessions
Lab rules
- Always use the distilled water from the green tap
- Always label things at the lab with the red tape. Put the initials, the lab name (PA), date and its contents
- For anything which needs to be autoclaved, label, stick the autoclave tape, and send for autoclaving
- Clean the bench before and after the work with ethanol
- Dry the cases before using them
- Keep a bottle/beaker of distilled water to use on the table
- No actual work is allowed after 5 or 6 pm - if the autoclaver is needed, the experiments requiring it should be presented to the operator by 4pm
The first week initial planning, 30 June - 3 July
- Tuesday
- Find the strain
- If plates are unavailable, the steps for preparing plates are:
- Cool the media to approximately 50 to 60 degree Celsius
- Fill the plate half full
- Dry them
- Get the strain from the freezer
- Streak the plates
- Incubate the plates
- Prepare other solution and medias needed
- Wednesday
- From the plates which were streaked and incubated on Tuesday, pick up a single colony using a loop
- Note: When looping, flame the loop till it is cherry red to reduce the risk of contamination
- Inoculate the colony with 3ml of LB
- Incubate the media in the incubator shaker at 37 degree Celsius to be used on Thursday
- Note: The tubes containing the media should be tilted at an angle for optimal growth conditions
- Thursday
- Prewarm the media at 37 degree Celsius
- Sample->wash 2x-? Suspend in 1ml->freeze->-80 degree Celsius
- Friday
- Transformation test with a plasmid
Lab session: 30th June
- We got samples of E.coli DH2 Alpha strains and placed them into three plates. They are placed for incubation at 37C
- Glycerol concentration will need be chanegd as 50%
- We have created 0.1M of CaCl2 using 5.5 gram of CaCl2. The final solution is 0.5 liter. We placed them into 4 boxes (three 100ml and one 200ml). They have blue covers. We have left them for autoclaving.
- We prepared LB solution using 20gr LB. Th final volume is 1 liter. We put them into boxes and left for autoclaving.
Lab session: 1st July
Things we need for today:
- LB
- Containers
- Sterile tubes
- Space in incubator for tonight
- We plated out single cultures from yesterday's plates for a practice.
- We inoculated 2ml of LB with single cultures from yesterday's plates and put them in the shaking incubator.
- We made up some 50% glycerol and it was sent to be autoclaved.
Things we need for tomorrow (Thursday):
- Have we used up a lot of anything- is there enough for Thursday?
- Sterile tubes
- Pipette tips
- Solutions
Culture
- Sterile flasks for culture (500ml)
- Spectrophotometer and cuvettes (where)
- Arrange space in shaking incubator
Harvest cells
- Centrifuge/ centrifuge vial (200ml?)
- Ice bucket and ice
- Glycerol
- Sterile microfuge tubes
- Liquid nitrogen
Lab session: 2nd July
- 1ml of preculture (which had been left overnight) was taken and used to inoculate 200ml of LB (standard bacterial broth). The flask which contained the mixture was then placed into the shaker, which both incubated the culture at 37 degrees Celsius and constantly shook the flask.
- Once 2 hours had elapsed, the optical densities of the cultures were taken using a spectrophotometer. This was to determine whether the cells had grown and if they had, where the cells were in their growth phase. For competency, E.coli need to be in early log phase. The results were as follows:
| Sample | Absorbance (nm) |
|---|---|
| 1 | 0.346 |
| 2 | 0.443 |
| 3 | 0.450 |
- The next step was to transfer the three cultures into a centrifuge and allow them to spin for 10 minutes, at a speed of 8,000 rpm and at a temperature of 4 degrees celsius. Because balance is required in a centrifuge (i.e. even numbers), we only used two of the tubes.
- Once spun, the supernatant was removed and the pellet was resuspended in calcium chloride, which had been placed in a fridge since the solution was made. The concentration of the calcium chloride was 0.1M and the volume was 40ml.
- The cells emersed in the calcium chloride were then held in ice for 40 minutes.
- Once 40 minutes was complete, the cells were once again centrifuged at a speed of 8,000 rpm and at the temperature of 4 degrees celsius. After being spun, the supernatant was removed and the pellet was resuspended in 1ml calcium chloride again. This solution was again pre-chilled in the fridge, was again 0.1M but only 1ml was applied this time round.
- 50% glycerol was then applied to the mixture. Because the concentration of the glycerol to cell mixture was supposed to be 10%, we applied 0.ml to 0.9ml of cell mixture.
- The resulting mixture was then placed into 50 microfuge tubes (each with a capacity of 100 microlitres) with these tubes being placed into a flask of liquid nitrogen. This process shock-freezes the E.coli cells.
- After being given a few minutes to confirm freezing of the cells, the tubes were immediately scooped out of the flask and transferred immediately into a -80 degrees celsius freezer for storage.
Lab session: 3rd July
Today we were transforming DNA using Phil's protocol, however we used the following changes:
- We added 3µl of DNA to our cells.
- We plated out 100µl of our dilutions.
- We incubated our transformed cells for the full 60 mins.
- We used the ampicillin resistance plasmid '435'
Procedure
- We followed the protocol using sterile technique where possible, as well as wearing gloves to prevent contamination of the DNA.
- Glass tops such as LB jars can be run under the flame, however plastic eppendorfs cannot so working around the flame is sufficient.
- We found it easier if the person pipetting held the eppendorf as they can see what they're doing better.
- Prof. Aldridges lab uses sterile beads to plate ourt rather than plastic spreaders, these are a bit fiddly so it may take a while to get used to tipping the right amount out!!
Our serial dilutions were prepared as follows:
- Using the waiting times within our procedure we set up 6 labelled eppendorf tubes with 900µl of LB, ready for the series dilution.
- We took 100µl of our incubated cells and put them in tube 1, mixed the solution briefly and then proceded to take 100µl of the diluted cells and tranfer them along the line of eppendorfs, resulting in eppendorfs containing 10-1 to 10-5 solutions for both the control and 'transformed' cells.
- We plated all 6 transformed E.coli solutions onto LB+amp agar plates, and the controls we plated onto both LB+amp plates(to check for contamination) as well as plain LB plates (in order to later calculate transformation frequency).
Lab Sessions - Week 2 (6th July - 10th July)
Lab session: 6th July
List of bricks for the day
- [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] (plasmid:pSB1AK3). Location: Plate 1, Well 13A
- [http://partsregistry.org/Part:BBa_I13522 BBa_I13522] (plasmid:pSB1AK3). Location: Plate 2, Well 8A
- [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] (plasmid:pSB1AT3). Location: Plate 1, Well 15A
Preparation of the tubes
- We got 3 tubes containing cells from the freezer at 80C and placed them into the ice and waited for 30 minutes
- We prepared 6 tubes (3 tubes for controls and 3 tubes for the plasmids) for the bricks
- We added 40ul of cells to each of the six tubes
- We added 3ul of DNA for the plasmid tubes and 3ul of water for the control tubes
- We added 900ul of LB for each of the six tubes
- We then placed the tubes in the shaking incubator for an hour.
Plating out
We prepared 9 plates with full strength (undiluted) and 9 plates with diluted tube content.
- Plates with undiluted content
- The first three plates having LB + Amp were used as the negative control. Hence the cells not having amp. resistance would not grow on these plates.
- The next three plates with LB content were used as the positive control. Cells without amp. resistanec would still grow on these plates.
- The other three plates were used for the plasmids.
| LB + Amp, (-) control 1 | LB, (+) control 1 | LB + Amp, Transformation Plasmid 1 |
| LB + Amp, (-) control 2 | LB, (+) control 2 | LB + Amp, Transformation Plasmid 2 |
| LB + Amp, (-) control 3 | LB, (+) control 3 | LB + Amp, Transformation Plasmid 3 |
- Plates with diluted content
- For the diluted ones we used 900ul of LB and 100ul of DNA from the corresponding tubes.
- The same set of 9 plates were prepared for the diluted content.
Lab Session:7th July
Experiment summary so far...
Yesterday, we attempted to transform cultures with BioBricks. We inserted the three BioBricks into competent E.coli cells grown in the previous lab session (via plasmids) and grew them overnight. The cells were spread on plates in both undiluted and diluted forms. The diluted solution of competent E.coli cells were spread onto three different plates - a negative control plate, a positive control plate and a plasmid plate (see table above). Today we reviewed the cultures which grew on the plates and prepared them for plasmid extraction and purification - this will form tomorrow's experiment.
Observations
Firstly, none of the E.coli cells mixed with BioBrick 1 (in the presence of competence inducers) took up the DNA - they didn't appear on the diluted or undiluted plates, which were lined with ampicillin. However the E.coli cells which had been mixed with BioBricks 2 and 3 did take up the DNA as they were present in colonies on ampicillin-covered plates. There were two colonies of bacteria which had been transformed by BioBrick 2, and eight colonies transformed by BioBrick 3. These 10 cultures became the focus of the subsequent experiments.
Preparation of Solutions
To prove that the DNA the E.coli cells had taken up really was the BioBrick added to the solution, we need to carry out the procedure of DNA electrophoresis with the aid of restriction enzymes. In order to carry out this step, we need to extract and purify the plasmids first (to be done tomorrow) via alkaline lysis - this requires three solutions, two of which were made today.
Solution 1
According to Dr. Aldridge's protocol named "Phil’s mini method for Alkaline Lysis for Mini Prep", we need to add 'solution 1' (along with an RNAse) to resuspended transformed E.coli cells, tomorrow. Solution 1 requires the following ingredients at the following concentrations:
* 50 mM Glucose * 25 mM Tris.Hcl (pH 8.0) * 10 mM EDTA (pH 8.0)
This solution was made today in preparation for tomorrow's exercise. We were told that the total volume should be 250ml. We were presented with pre-made solutions - 1M Tris HCl and 0.5M EDTA - and were told we needed to calculate the correct volumes of each of these solutions to be added to the beaker to give the desired molarity.
We were presented with glucose in it's solid form and asked to make a stock solution of 1M glucose; after which we were asked to calculate the amount to glucose solution to add to the beaker to make a molarity of 0.5M! The following equation helped:
Molecular Weight x Desired Volume (Litres) x Molarity = Weight (grams)
- Making the glucose stock solution
- molecular weight of glucose = 180.2
- desired volume of stock solution = 500ml
- desired molarity of stock = 1M
Put these values into the equation above gives the amount of glucose solid needed for 500ml of water...
180.2 x 0.5l x 1M = 90.1g
We added to 90.1 grams of glucose to 0.5 litres to get a 1M stock solution.
- Adding the solutions to the beaker
- 25ml of 1M glucose stock solution was transferred to beaker (to make 50mM)
- 6.25ml of 1M Tris.HCl was transferred to the same beaker (to make 25mM)
- 5ml of 0.5M EDTA also added (to make 10mM)
- H2O made the rest of the solution
These values are derived from the above equation
Solution 3
We also needed to make solution 3 for tomorrow's practical. This involved the following ingredients:
* 5M Potassium acetate: 60ml * glacial acetic acid: 11.5 ml * H2O: 28.5 ml
The glacial acetic acid and the water could be added to the 'Solution 3' beaker immediately, but calculations had to be carried out for the 5M potassium acetate to determine the mass of the solid. We decided to make a 200ml stock of 5M solution.
- The molecular weight of potassium acetate = 98.15
- The desired volume of this solution = 0.2 litres
- The desired molarity = 5M
Put this into the equation:
Molecular Weight x Desired Volume (Litres) x Molarity = Weight (grams)
98.15 x 0.2l x 5M = 98.15 grams
We then made up the solution and added it to the other ingredients, according to the protocol.
Growing the transformed cultures
With the solutions prepared and sent to the autoclaver, we had the task of taking each of the 10 colonies of transformed cells and placing them into 10 tubes containing LB. We labelled the tubes 1-10 and added the colonies to the tubes as follows:
- Tube 1 = BioBrick 2 - colony 1
- Tube 2 = BioBrick 2 - colony 2
- Tube 3 = BioBrick 3 - colony 1
- Tube 4 = BioBrick 3 - colony 2
- Tube 5 = BioBrick 3 - colony 3
- Tube 6 = BioBrick 3 - colony 4
- Tube 7 = BioBrick 3 - colony 5
- Tube 8 = BioBrick 3 - colony 6
- Tube 9 = BioBrick 3 - colony 7
- Tube 10 = BioBrick 3 - colony 8
- BioBrick 2 = [http://partsregistry.org/Part:BBa_I13522 BBa_I13522] in the pSB1AK3 plasmid
- BioBrick 3 = [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] in the pSB1AT3 plasmid
We then left the cultures to grow overnight.
Lab session: 8th July
Experiment Recap
In summary, our team have attempted to transform E.coli with three BioBricks - [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] (with plasmid pSB1AK3), [http://partsregistry.org/Part:BBa_I13522 BBa_I13522] (with plasmid pSB1AK3) and [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] (with plasmid PSB1AT3). The cells were cultured on ampicillin-covered agar plates overnight after being treated with both the BioBrick DNA and the competence inducer calcium chloride. Only 10 colonies emerged on the ampicillin-covered plates - 2 cultures transformed with BioBrick BBa_I13522 and 8 cultures transformed with BioBrick BBa_J04450.
These cultures were then placed in some nutrient broth and left overnight to replicate and amplify the plasmid concentration. Today, we will process these cultures of cells by extracting and purifying the plasmid DNA - this will then be used in DNA electrophoresis to confirm that the DNA inherited by the ampicillin resistant bacteria are the BioBricks we inserted.
Observations

The above image shows the appearance of the 10 tubes after a night of incubation in the shaker. Tubes 1 and 2 showed yellow, translucent cultures, whereas tubes 3-10 showed red, translucent cultures. This is because tubes 1 and 2 contain E.coli transformed by the biobrick [http://partsregistry.org/Part:BBa_I13522 BBa_I13522] (with plasmid pSB1AK3) which encodes Green Fluorescence Protein (GFP) whereas tubes 3-10 carry the BioBrick [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] (with plasmid PSB1AT3), which encodes RFP (Red Fluorescence Protein).
Today's Procedure
- spin and resuspend the 10 cultures in 300ul of Sol.I+RNase
- Add 600 ul fresh Sol.II
- 5 min RT then add 250 ul Sol III
- Centrifuge for 20 mins
- Put 1ml supernatant into new tube and add 600 ul Isopropanol
- Spin for 15 min and aspirate
- Add 500 ul 70% ethanol and spin for 5 min
- Aspirate and speed vac resuspending in 50 ul water
Making up Solution II
The procedure states that Solution II should be made 'fresh' so we had to make it today and before we carried out the rest of the procedure. The solution is made up of the following:
- 2ml NaOH (1M)
- 1ml 10%SDS
- 7 ml water
Because the volumes and the molarities of the recipe were given and because the solutions of NaOH and SDS were made up already, there was no need for calculations.
Carrying out the rest of the procedure
- The procedure demanded us to centrifuge 3ml (at least) of the 10 cultures. However the Eppendorf tubes that are placed into the centrifuge dont't have that capacity. So we pipetted 1.5ml of the 10 cultures into 10 Eppendorf tubes, spun them, removed the supernatant and added a further 1.5ml of the cultures to the 10 tubes. This was spun and the supernatant removed.
- The 10 pellets were then each resuspended in 300 microlitres of Solution I. Prior to this, we added 150 microlitres of RNAse A to the bottle containing our premade Solution I.
- 600 microlitres of the newly made Solution II was added to each of the 10 tubes. The tubes were then left (sealed, of course) in the rack for 5 mins.
- 250 microlitres of Solution III was added to each of the 10 tubes and the resulting solution centrifuged for 20 minutes. The resulting solution could be seen as a cloudy white precipitate of chromosomal DNA and protein. Instead of removing the supernatant and disposing of it, once the spinning had stopped we extracted 1ml of supernatant and placed them into 10 new Eppendorf Tubes (labelled '1S', '2S', '3S',...etc). The tubes containing the pellets were then discarded.
- To the 10 tubes containing supernatant, 600 microlitres of prepared isopropanol was added. These tubes were then placed in the centrifuge for a further 15 mins.
- The resulting supernatants for the 10 tubes were then removed and any droplets of solution were carefully removed by a vacuum pump (without disrupting the DNA which had gathered on one side of the Eppendorf tubes). To this, 500 microlitres of ethanol (at 70%) was added.
- Making 70% ethanol:
- 35 ml of ethanol added to beaker
- 15 ml of water added to beaker
- The resulting solution was then spun in the centrifuge for 5 minutes.
- The supernatant was removed and the remaining droplets of solution removed without disrupting the transparent pellet the plasmids had formed. Once the plasmid pellet was dried of all the solution, it was resuspended in 50 microlitres of water. The tubes were then lightly centrifuged (for a few seconds) to make sure all of the water, along with the DNA, had congregated at the bottom of the tubes.
Conclusion
We are now ready to carry out DNA electrophoresis on the 10 samples, which will happen tomorrow.
Lab session: 9th July
Experiment Recap
Up to this point, we have successfully isolated and cultured single colonies of E.coli cells. With these micro-organisms, we were able to induce competence and had the opportunity to transform them with the following BioBricks:
- [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] with plasmid pSB1AK3
- [http://partsregistry.org/Part:BBa_I13522 BBa_I13522] with plasmid pSB1AK3
- [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] with plasmid pSB1AT3
So far, we are convinced that 2 colonies of our cultured E.coli cells have become transformed by BioBrick BBa_I13522 (with plasmid pSB1AK3) which encodes GFP and that 10 colonies of our cultured E.coli cells have been transformed by BioBrick BBa_J04450 (with plasmid pSB1AT3), which leads to the production of RFP. This can be seen visually in the tubes; see image in the previous experiment under the heading 'Observations'.
However visual representations may not always available and in these scenarios, it is difficult to tell whether the E.coli have become transformed by the given BioBrick unless DNA analysis is conducted. Even though the cells have survived the negative selection process (i.e. antibiotic exposure), it doesn't mean that they all possess the BioBrick - they could have received that resistance through other means.
Aim of Today's Experiment
We need to get ourselves familiarised with conducting DNA gel electrophoresis. We shall be doing this by preparing and then inserting the plasmid DNA (extracted from the 10 colonies of cells) into the gel. At this stage, the plasmids will be inserted as uncut objects because no restriction enzymes have been added yet (the analysis of cut plasmids comes at a later stage). Thus we won't be proving that it is the BioBrick which is causing the bacteria to display the properties seen so far. We will be proving that the bacteria have taken up plasmids.
This procedure will see the team:
- Prepare the DNA electrophoresis equipment by preparing the gel, administering buffer and ethidium bromide, etc
- Prepare the plasmid DNA for gel electrophoresis.
- Apply the DNA to the gel and conduct electrophoresis
- Capture an image of the gel using UV-scanner.
Experiment procedures
The protocol we used for this practical can be found 'here.'
Preparing the gel
- A premade stock solution of agarose gel (0.8% agarose) was taken and heated in microwave for 3 minutes under medium/high power initially. Short 30 second bursts were required to completely liquify the solution
- Gel was then removed from microwave and left on bench to cool
Preparing the buffer
- Premade solution of 1x TAE was used - took volume of this and added to flask
- Added 8 microlitres to the TAE buffer
Preparing the gel electrophoresis equipment
- Trays and comb were cleaned and provided for the team, thanks to the lab staff!
- Inserted comb into groove nearest the edge of the tray and placed in proximity of the electrodes.
Pouring Buffer and Gel
- Placed the 1x TAE buffer (plus ethidium bromide) into the tray (with gel comb inserted in groove nearest negative electrode)
- Agarose gel solution placed into tray and bubbles removed using a pipette. After this, lid placed over the gel electrophoresis equipment.
Preparing DNA samples
- Removed 10 Eppendorf tubes (containing the plasmid DNA for the 10 colonies of E.coli) from the freezer and defrosted them briefly. In the meantime, 10 fresh Eppendorf tubes were placed in rack and labelled '1-10' to match the 10 plasmid DNA Eppendorf tubes.
- When defrosted, 9 microlitres of the plasmid DNA was transferred to new Eppendorf tubes. 1 microlitre of dye was then added to the new Eppendorf tubes. The tubes were then lightly centrifuged to make sure the dye and the plasmid DNA had mixed at the bottom of the tube.
- An Eppendorf tube containing the standard DNA ladder and dye was added to the rack, in the first entry of the row with the 'DNA + dye' tubes following afterwards (in order of 1-10). This was then taken over to the gel electrophoresis tray.
Loading the samples and carrying out electrophoresis
- Made sure agarose gel had completely set. After confirmation of setting, the gel comb was removed revealing wells in which to put the DNA into.
- Making sure that the pipette was pointed vertically over the well and that the pipette was hovering over (and not poking) the well, 10 microlitres of DNA was slowly and gently added to each well.
- Well 1 = DNA Ladder
- Well 2 = plasmid DNA from colony 1 - BBa_I13522 with plasmid pSB1AK3
- Well 3 = plasmid DNA from colony 2 - BBa_I13522 with plasmid pSB1AK3
- Well 4 = plasmid DNA from colony 3 - BBa_J04450 with plasmid pSB1AT3
- Well 5 = plasmid DNA from colony 4 - BBa_J04450 with plasmid pSB1AT3
- Well 6 = plasmid DNA from colony 5 - BBa_J04450 with plasmid pSB1AT3
- Well 7 = plasmid DNA from colony 6 - BBa_J04450 with plasmid pSB1AT3
- Well 8 = plasmid DNA from colony 7 - BBa_J04450 with plasmid pSB1AT3
- Well 9 = plasmid DNA from colony 8 - BBa_J04450 with plasmid pSB1AT3
- Well 10 = plasmid DNA from colony 9 - BBa_J04450 with plasmid pSB1AT3
- Well 11 = plasmid DNA from colony 10 - BBa_J04450 with plasmid pSB1AT3
- Lid was applied and electrodes positioned correctly (positive electrode at end furthest from wells and negative electrode positioned nearest wells). Power was switched on and voltage set to 150 volts. This process was allowed to run for 40 minutes.
Observations and Results
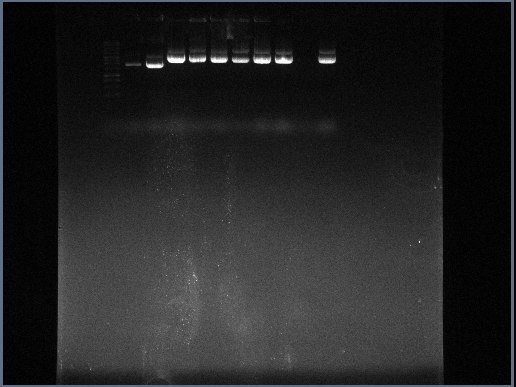
- Once 40 minutes had expired, electrodes were switched off and lid removed. In the gel it could be seen that the DNA had streaked from the wells to the positive electrode. Three marker bands were visible, in order from nearest wells to nearest positive electrode - a cyan band, a dark blue band and a yellow band.
- Gel removed from gel tray and taken to UV scanner. The following photograph was produced:
From this photograph it can be seen that apart from the E.coli cells located in colony 9, all the colonies that had survived the negative selection and displayed either a red or a green colour in solution contained plasmid DNA. This proves that most of the colonies had been transformed with a plasmid at least. The possibilities for colony 9 not revealing a plasmid DNA readout are as follows:
- Colony 9 E.coli cells did not receive the plasmid DNA and acquired it's ampicillin resistance and colour properties elsewhere
- The plasmid DNA (which appears as a transparent pellet in the previous experiment) was lost when the supernatant was tipped out.
Now that Team:Newcastle has proved that the competent E.coli cells have taken up the plasmids, the next step will be to prove that the 'E.coli' have taken up the specific BioBricks that we attempted to transform them with. This will involve restriction enzymes and another DNA gel electrophoresis.
News
Events
- 20 – 21 June 2009 - Europe workshop (London)
- 23 – 24 June 2009 - UK iGEM meetup (Edinburgh)
- 23 October Practice Presentation (Newcastle)
- 23 October T-shirts are ready
- 27 October Practice Presentation (Sunderland)
- 27 October Poster is ready
- 30 October – 2 November 2009 - Jamboree (Boston)
Social Net
- Newcastle iGEM Twitter
- [http://www.facebook.com/home.php#/group.php?gid=131709337641 Newcastle on Facebook]
- [http://www.youtube.com/user/newcastle2009igem Newcastle Youtube Channel]
 "
"