Team:Newcastle/Promoter Library
From 2009.igem.org
(→Promoter Library) |
(→Promoter Library) |
||
| Line 6: | Line 6: | ||
Please click on the following links to learn more about the promoter library sub-project: | Please click on the following links to learn more about the promoter library sub-project: | ||
<br> | <br> | ||
| + | *[[#Introduction|Introduction]] | ||
| + | *[[#Novelty in this sub-project|Novelty in this sub-project]] | ||
| + | *[[#Design|Design]] | ||
| + | *[[#Lab Work Strategies|Lab Work Strategies]] | ||
| + | *[[#Progress|Progress]] | ||
<br> | <br> | ||
==Introduction== | ==Introduction== | ||
Revision as of 02:22, 22 October 2009
Promoter Library
Please click on the following links to learn more about the promoter library sub-project:
Introduction
In order to effectively model genetic circuits, databases of parts with known characteristics are essential. Without such databases, computationally designed circuits may not be feasible to implement in vivo. Unfortunately, parameters such as promoter strength, RBS affinity, transcription, translation and decay rates are hard to find in the literature, and those which do exist are rarely measured under standardised conditions.
Several efforts to produce parts libraries are currently underway. As part of our contribution to synthetic biology, we decided to create a promoter library for B. subtilis.
Novelty in this sub-project
The novelty in this sub-project lies in the generation of a large number of B. subtilis promoters, based upon carefully considered variations to an existing promoter region. We chose the promoter region of the transcription factor SigmaA, since it is the most widely-used promoter in B. subtilis. Over 300 genes are controlled by SigA. Their promoter regions have highly conserved -10 and -35 regions, surrounded by variable sequences.
Design
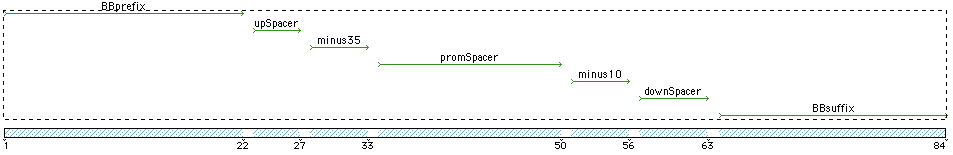
Template used to create the promoter sequences:
Prefix...UpSpacer...-35...PromSpacer...-10...DownSpacer..Suffix
- BBPrefix - StandardBioBrickPrefix EcoR1 and Xba1 - gaattcgcggccgcttctagag
- UpSpacer - attta - consensus sigA 5 bases 5'
- -35 - TTGACA
- PromSpacer - length 17 - consensus ttttatttaaattatga
- -10 - TATAAT
- DownSpacer (from ackA)- ggaaaag
- BBSuffix - Standard BioBrick suffix Spe1 and Pst1 - tactagtagcggccgctgcag
Variants
We designed three degenerate sequences to create a library of promoters which we hope will have different strengths.
Variant 1
In this variant we only modified the -35 and -10 consensus sequences.
BBPrefix...UpSpacer...N-35... PromSpacer...N-10...DownSpacer..BBSuffix
Variant 2
In the second variant we modified the promoter sequence between the -35 and -10 regions.
BBPrefix...UpSpacer...-35... 18N’s...-10...DownSpacer..BBSuffix
Variant 3
In the third variant we changed the last two bases of the minus 35 region and the first two bases of the -10 region.
BBPrefix...UpSpacerNN...-35...NNPromSpacerNN...-10...NNDownSpacer..BBSuffix
Lab Work Strategies
- Synthesis other strand using polymerase and a primer
- Cut promoter fragments using EcoR1 and Pst1
- Cut pSB1AT3 vector using EcoR1 and Pst1
- Ligate the insert and the plasmid backbone, and transform E. coli DH5alpha
- Subclone into Bacillus integration vector with gfp and insert at amyE
Progress
The consensus and variant sequences were designed and ordered from GeneArt. Once received they were PCRd, but we ran out of time, and the library has not been characterised.
News
Events
- 20 – 21 June 2009 - Europe workshop (London)
- 23 – 24 June 2009 - UK iGEM meetup (Edinburgh)
- 23 October Practice Presentation (Newcastle)
- 23 October T-shirts are ready
- 27 October Practice Presentation (Sunderland)
- 27 October Poster is ready
- 30 October – 2 November 2009 - Jamboree (Boston)
Social Net
- Newcastle iGEM Twitter
- [http://www.facebook.com/home.php#/group.php?gid=131709337641 Newcastle on Facebook]
- [http://www.youtube.com/user/newcastle2009igem Newcastle Youtube Channel]
 "
"