Team:Newcastle/Stochasticity
From 2009.igem.org
(→BioBrick constructs) |
(→Introduction) |
||
| (22 intermediate revisions not shown) | |||
| Line 7: | Line 7: | ||
==Introduction== | ==Introduction== | ||
| - | One of the most | + | One of the most exciting aspects of our project is our synthetic stochastic switch. The switch regulates the decision to become a non-germinating metal container spore, or a spore that can go on to germinate as part of the normal life cycle. Whilst stochastic oscillators have been implemented before using transcriptional regulators, our switch makes use of an invertable DNA segment to ensure that the decision is heritable. |
| + | |||
| + | By differentially controlling the expression of the Hin invertase, we designed our switch to be tunable to achieve a biased heads or tails response, allowing a range of probabilities of orientation of the invertable segment to be achieved. | ||
| + | |||
| + | [[Image:Newcastle r Switchv2.gif|center]] | ||
==Novelty in this sub-project== | ==Novelty in this sub-project== | ||
| - | We | + | We designed a synthetic stochastic switch by using an invertible segment of DNA flanked by a pair of promoters. Depending on the orientation of the invertible sequence, coding sequences will be expressed which reflect the decision to be a metal container or not. We also tuned the natural stochasticity of the sporulation system towards greater sporulation rates by altering the rate of ''Spo0A'' phosphorylation. |
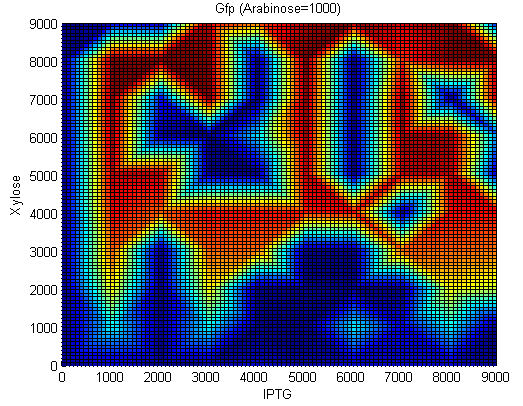
[[Image:Team_Newcastle_iGEM_2009_StochasticSwitch_GFP_2.png|thumb|center|350px|Gfp concentrations. IPTG:[0-9000nM], Xylose=[0-9000nM], Arabinose=1000nM]] | [[Image:Team_Newcastle_iGEM_2009_StochasticSwitch_GFP_2.png|thumb|center|350px|Gfp concentrations. IPTG:[0-9000nM], Xylose=[0-9000nM], Arabinose=1000nM]] | ||
| Line 17: | Line 21: | ||
==BioBrick constructs== | ==BioBrick constructs== | ||
| - | There | + | There are a number of bricks involved within the stochastic switch construct. |
| - | The stochastic brick construct uses the Hin invertase system in order to flip a | + | |
| + | The stochastic brick construct uses the Hin invertase system in order to flip a region between Hix sites. The directionality of the promoter determines whether the switch is 'on' or 'off'. When the promoter is facing right it allows transcription of genes that control: | ||
| + | |||
** Prevention of germination | ** Prevention of germination | ||
** Upregulation of sporulation rate | ** Upregulation of sporulation rate | ||
| Line 24: | Line 30: | ||
** Decreased cadmium efflux | ** Decreased cadmium efflux | ||
** Upregulation of cadmium import | ** Upregulation of cadmium import | ||
| + | |||
| + | |||
| + | Importantly, Hin is '''differentially expressed''' depending on the levels of the two inducible promoters that flank the invertable segment on which it lies. This means the segment can be '''biased''' in a predictable and controllable fashion to favour one orientation or the other. | ||
| + | |||
The following diagram shows our stochastic construct: | The following diagram shows our stochastic construct: | ||
| Line 29: | Line 39: | ||
[[Image:Team NewcastleStochastic switch.png| center|550px]] | [[Image:Team NewcastleStochastic switch.png| center|550px]] | ||
| - | [[Image:Team newc Stoch key.png | + | [[Image:Team newc Stoch key.png| 200px]] |
===Prevention of germination=== | ===Prevention of germination=== | ||
| - | The prevention of germination is governed by another invertase switch. | + | The prevention of germination is governed by another invertase switch. When the sequence faces right, a FimE protein is expressed which inverts a further promoter region. This promoter controls expression of the ''cwlD'' |
| - | and '' | + | and ''sleB'' genes. If their promoter is in the correct orientation then the cell will be able to germinate and continue as a vegetative cell. However if their promoter has been flipped, the cell can not germinate following sporulation, and will be trapped as a metal containing spore. |
===Upregulation of sporulation rate=== | ===Upregulation of sporulation rate=== | ||
The upregulation of sporulation involves increasing KinA expression. ''kinA'' codes a kinase protein that phosphorylates the Spo0A protein to its active form. When the promoter region within our stochastic brick faces right, there will be increased KinA expression, and thus a greater sporulation rate. | The upregulation of sporulation involves increasing KinA expression. ''kinA'' codes a kinase protein that phosphorylates the Spo0A protein to its active form. When the promoter region within our stochastic brick faces right, there will be increased KinA expression, and thus a greater sporulation rate. | ||
| - | ===Metal sponge and cadmium influx/efflux=== | + | ===Metal sponge and cadmium influx/efflux=== |
| - | Expression of the metallothionein fusion protein ('' | + | Our stochastic switch determines whether the spores can germinate, or whether they are commited to be metal containers that cannot germinate again. We need this switch as we cannot totally interrupt the natural life cycle of the bacteria, since a proportion of cells have to go on to seed the next generation. |
| + | Expression of the metallothionein fusion protein (''cotC-gfp-smtA''), cadmium import channel (''mntH'') and the cadmium efflux channel (''cadA'') is also governed by the direction of the stochastic promoter. When the direction of promoter faces right, the metallothionein fusion protein's expression will be triggered, ant will soak up the cadmium. While the import channel is upregulated, the efflux system's activity will be slowed down to increase the amountof cadmium inside the cell. | ||
===Stochastic Brick=== | ===Stochastic Brick=== | ||
| - | We | + | We decided to get our stochastic construct synthesised, as trying to build the construct manually would be too time consuming. The following sequencher diagram shows the components of the construct we had synthesised. |
[[Image:Team newc Sequencher synth stoch.png| center|600px]] | [[Image:Team newc Sequencher synth stoch.png| center|600px]] | ||
| - | |||
| - | |||
| - | |||
| - | |||
===Testing construct=== | ===Testing construct=== | ||
| - | In order to test our construct we | + | In order to test our construct we had to redesign using inducible promoters governing Hin invertase expression. We used the promoters ''pSpac'' and ''pxylA'' (Induced by IPTG and Xylose) to test our system. We include cut sites around these promoters in order to replace them with SigmaA promoters once the construct has been characterised.(See sequencher diagram above) |
===Degradation controller=== | ===Degradation controller=== | ||
| - | In order to have another level of control over the orientation of the promoter within the flipping region we | + | In order to have another level of control over the orientation of the promoter within the flipping region we added a degradation tag to the Hin invertase protein. The following paper describes how proteins including modified ''ssrA'' tags can be located to the ClpXP protease by an Sspb protein. This means that inducible Sspb expression can requlate degradation levels of the tagged protein. |
| - | [http://www3.interscience.wiley.com/journal/121415079/abstract?CRETRY=1&SRETRY=0 Inducible protein degradation in Bacillus subtilis using heterologous peptide tags and adaptor proteins to target substrates to the protease ClpXP ] | + | [http://www3.interscience.wiley.com/journal/121415079/abstract?CRETRY=1&SRETRY=0 Inducible protein degradation in ''Bacillus subtilis'' using heterologous peptide tags and adaptor proteins to target substrates to the protease ClpXP ] |
| - | We | + | We decided to put the Sspb protein under the control of an arabinose inducible promoter as the following diagram illustrates. We also included a region of the ''sac'' gene in our construct, so that the region will integrate into the ''Bacillus'' genome at a region other than ''amyE''. |
[[Image:Team NewcIntegration Deg control.png |center|500px]] | [[Image:Team NewcIntegration Deg control.png |center|500px]] | ||
| - | We added a modified version of ''ssrA'' degradation tag onto the C-terminus of Hin protein. | + | We added a modified version of ''ssrA'' degradation tag onto the C-terminus of the Hin protein. Expressed proteins are therefore degraded by ClpXP. However mutations on the ''ssrA'' tag weaken the recognition by ClpX, and the modified tags require the SspB adaptor protein to be recognized. When the SspB protein is expressed the proteins tagged with modified version of ''ssrA'' tag are targeted for degradation. Otherwise they remain stable. |
| - | In ''B. subtilis'' there is no ''sspB'' orthologue and SspB from ''E. coli'' works in ''B. subtilis''. By regulating the levels of SspB by arabinose, we | + | In ''B. subtilis'' there is no ''sspB'' orthologue and SspB from ''E. coli'' works in ''B. subtilis''. By regulating the levels of SspB by arabinose, we implemented an inducable protein degradation device. |
| - | [[Image:Team_Newcastle_iGEM_2009_Degradation_Model_4.png|thumb|center|400px|Hin vs | + | [[Image:Team_Newcastle_iGEM_2009_Degradation_Model_4.png|thumb|center|400px|Hin vs SspB according to the speed of degradation by ClpXP]] |
| - | + | The wild type ''E. coli'' ''ssrA'' tag is '''AANDENY-ALAA''' (SspB recognition site – ClpX recognition site). As suggested in the paper, we took one of the modified ''ssrA'' tags to use in our system. | |
'''AANDENY-SENY-ALGG''' (SspB recognition site – SENY +4 Linker - ClpX recognition site) | '''AANDENY-SENY-ALGG''' (SspB recognition site – SENY +4 Linker - ClpX recognition site) | ||
| - | This tag works well in ''B. subtilis'' | + | This tag works well in ''B. subtilis''. However, degradation tags can affect the activity of proteins. Different degradation tags may effect the activity of different proteins. It has been shown that this tag effected the activity of ComA(1). |
| - | #Griffith, K. L., and A. D. Grossman. 2008. Inducible protein degradation in Bacillus subtilis using heterologous peptide tags and adaptor proteins to target substrates to the protease ClpXP. Mol. Microbiol. 70:1012-1025. | + | #Griffith, K. L., and A. D. Grossman. 2008. Inducible protein degradation in ''Bacillus subtilis'' using heterologous peptide tags and adaptor proteins to target substrates to the protease ClpXP. Mol. Microbiol. 70:1012-1025. |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
===Stochastic Modelling Tools=== | ===Stochastic Modelling Tools=== | ||
| - | '''Matlab''' can be used for stochastic modelling. Glasgow team used Matlab | + | '''Matlab''' can be used for stochastic modelling. The Glasgow team used Matlab to implement the Gillespie algorithm for incorporating noise among cells. They also used deterministic modelling using ODEs and compared their results. When the number of cells increase two approaches become similar since the noise is cancelled out. |
'''Stocks 2''' is another stochastic simulation tool which also uses Gillespie’s direct method and supports SBML. | '''Stocks 2''' is another stochastic simulation tool which also uses Gillespie’s direct method and supports SBML. | ||
| - | |||
| - | + | We used computational modelling in Matlab to try to determine how to make our system tuneable. | |
| - | + | ||
| - | We | + | |
Please see our [[Team:Newcastle/Modelling|modelling]] page for Matlab files on our stochastic switch model. | Please see our [[Team:Newcastle/Modelling|modelling]] page for Matlab files on our stochastic switch model. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
===FimE switch=== | ===FimE switch=== | ||
| - | The FimE switch is a similar switch to the Hix system, | + | The FimE switch is a similar switch to the Hix system. However, it acts as a latch, meaning that once flipped the segmant will not flip back. |
# [http://genomics.lbl.gov/Stuff/TimHam-BandB-online%20version.pdf fimE switch for DNA re-arrangement] | # [http://genomics.lbl.gov/Stuff/TimHam-BandB-online%20version.pdf fimE switch for DNA re-arrangement] | ||
A Tightly Regulated Inducible Expression System Utilising the fim Inversion Recombination Switch.(''E. Coli'') Timothy S. Ham, Sung Kuk Lee, Jay D. Keasling,Adam P. Arkin,Received 21 December 2005; accepted 2 March 2006 Published online 13 March 2006 in Wiley InterScience (www.interscience.wiley.com). DOI: 10.1002/bit.20916 | A Tightly Regulated Inducible Expression System Utilising the fim Inversion Recombination Switch.(''E. Coli'') Timothy S. Ham, Sung Kuk Lee, Jay D. Keasling,Adam P. Arkin,Received 21 December 2005; accepted 2 March 2006 Published online 13 March 2006 in Wiley InterScience (www.interscience.wiley.com). DOI: 10.1002/bit.20916 | ||
| Line 144: | Line 112: | ||
And to find out how we are tuning sporulation using our stochastic switch choice see the sporulation tuning page. | And to find out how we are tuning sporulation using our stochastic switch choice see the sporulation tuning page. | ||
| + | ===Lab strategies=== | ||
| + | To carry out our labwork we needed cloning strategies for all of our bricks and devices. Please see our [[Team:Newcastle/ Stochastic Switch cloning strategy| cloning strategies]] page for details on how we cloned our devices. | ||
| + | {|style="color:DarkBlue;background-color:#ffffcc;" cellpadding="20" cellspacing="0" border="1" | ||
| + | ! colspan="2" |<font size=3> <center>'''Summary of lab work success:'''</center></font> | ||
| + | |- | ||
| + | |'''Date:''' | ||
| + | |'''Achievement:''' | ||
| + | |- | ||
| + | |11/09/09 | ||
| + | |Successfully cloned the ''sspB'' degradation controller fragment into pSB1AT3 | ||
| + | [[Team:Newcastle/Labwork/11_September_2009 | Lab book]] | ||
| + | |- | ||
| + | |18/09/09 | ||
| + | |Sucessfully cloned the ''ara'' promoter/ operator fragment into pSB1AT3 | ||
| + | [[Team:Newcastle/Labwork/18_September_2009 | Lab book]] | ||
| + | |- | ||
| + | |24/09/09 | ||
| + | |Successfully cloned the ''sspB'' fragment into the ''ara'' + pSB1AT3 prepared backbone. We now have an arabinose inducible degradation controller! | ||
| + | [[Team:Newcastle/Labwork/24_September_2009| Lab book]] | ||
| + | |} | ||
{{:Team:Newcastle/Footer}} | {{:Team:Newcastle/Footer}} | ||
{{:Team:Newcastle/Right}} | {{:Team:Newcastle/Right}} | ||
Latest revision as of 02:23, 22 October 2009
Stochastic Switch
Introduction
One of the most exciting aspects of our project is our synthetic stochastic switch. The switch regulates the decision to become a non-germinating metal container spore, or a spore that can go on to germinate as part of the normal life cycle. Whilst stochastic oscillators have been implemented before using transcriptional regulators, our switch makes use of an invertable DNA segment to ensure that the decision is heritable.
By differentially controlling the expression of the Hin invertase, we designed our switch to be tunable to achieve a biased heads or tails response, allowing a range of probabilities of orientation of the invertable segment to be achieved.
Novelty in this sub-project
We designed a synthetic stochastic switch by using an invertible segment of DNA flanked by a pair of promoters. Depending on the orientation of the invertible sequence, coding sequences will be expressed which reflect the decision to be a metal container or not. We also tuned the natural stochasticity of the sporulation system towards greater sporulation rates by altering the rate of Spo0A phosphorylation.
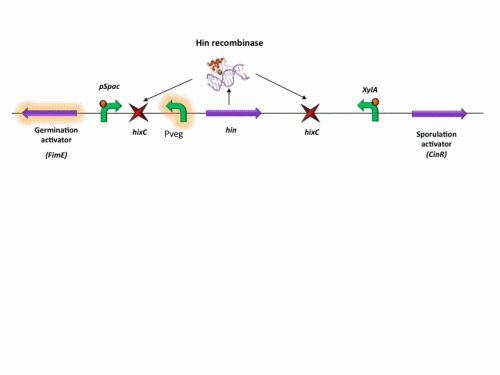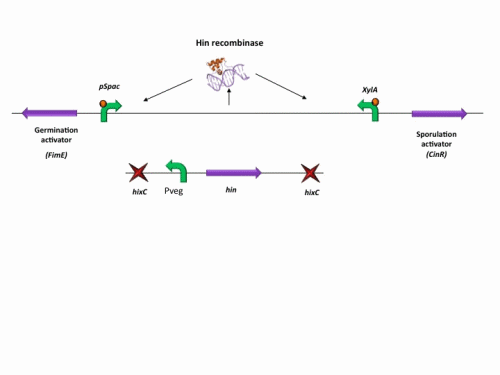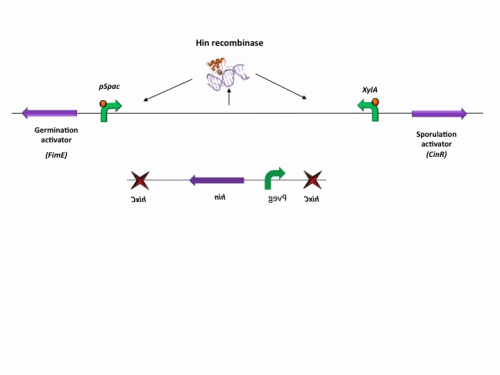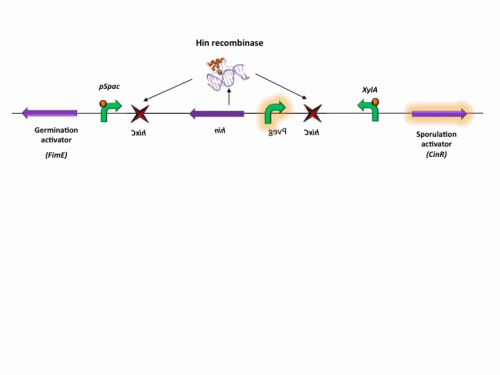
BioBrick constructs
There are a number of bricks involved within the stochastic switch construct.
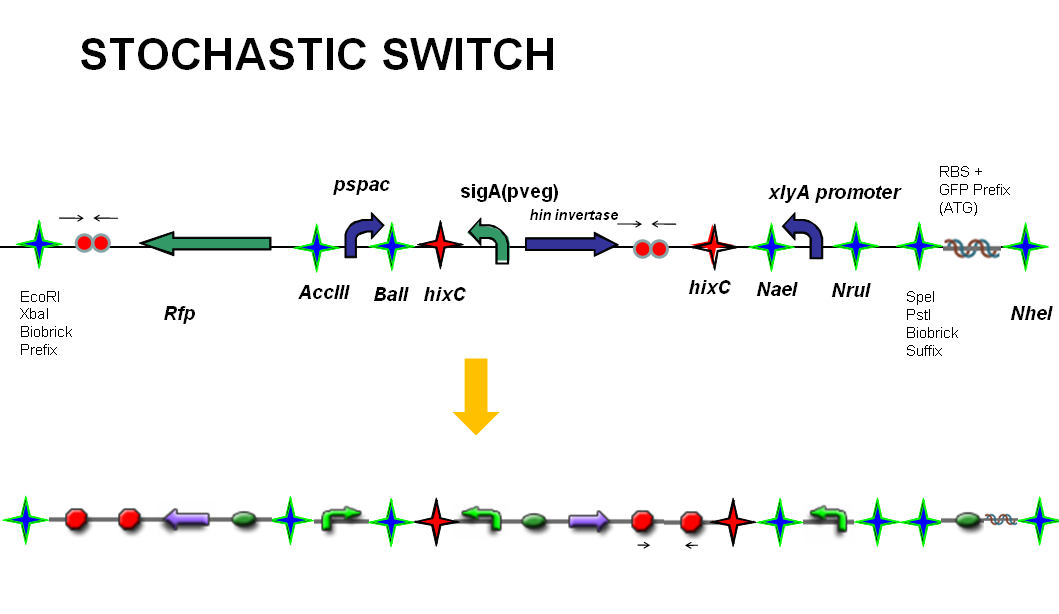
The stochastic brick construct uses the Hin invertase system in order to flip a region between Hix sites. The directionality of the promoter determines whether the switch is 'on' or 'off'. When the promoter is facing right it allows transcription of genes that control:
- Prevention of germination
- Upregulation of sporulation rate
- Expression of the metal sponge (SmtA)
- Decreased cadmium efflux
- Upregulation of cadmium import
Importantly, Hin is differentially expressed depending on the levels of the two inducible promoters that flank the invertable segment on which it lies. This means the segment can be biased in a predictable and controllable fashion to favour one orientation or the other.
The following diagram shows our stochastic construct:
Prevention of germination
The prevention of germination is governed by another invertase switch. When the sequence faces right, a FimE protein is expressed which inverts a further promoter region. This promoter controls expression of the cwlD and sleB genes. If their promoter is in the correct orientation then the cell will be able to germinate and continue as a vegetative cell. However if their promoter has been flipped, the cell can not germinate following sporulation, and will be trapped as a metal containing spore.
Upregulation of sporulation rate
The upregulation of sporulation involves increasing KinA expression. kinA codes a kinase protein that phosphorylates the Spo0A protein to its active form. When the promoter region within our stochastic brick faces right, there will be increased KinA expression, and thus a greater sporulation rate.
Metal sponge and cadmium influx/efflux
Our stochastic switch determines whether the spores can germinate, or whether they are commited to be metal containers that cannot germinate again. We need this switch as we cannot totally interrupt the natural life cycle of the bacteria, since a proportion of cells have to go on to seed the next generation. Expression of the metallothionein fusion protein (cotC-gfp-smtA), cadmium import channel (mntH) and the cadmium efflux channel (cadA) is also governed by the direction of the stochastic promoter. When the direction of promoter faces right, the metallothionein fusion protein's expression will be triggered, ant will soak up the cadmium. While the import channel is upregulated, the efflux system's activity will be slowed down to increase the amountof cadmium inside the cell.
Stochastic Brick
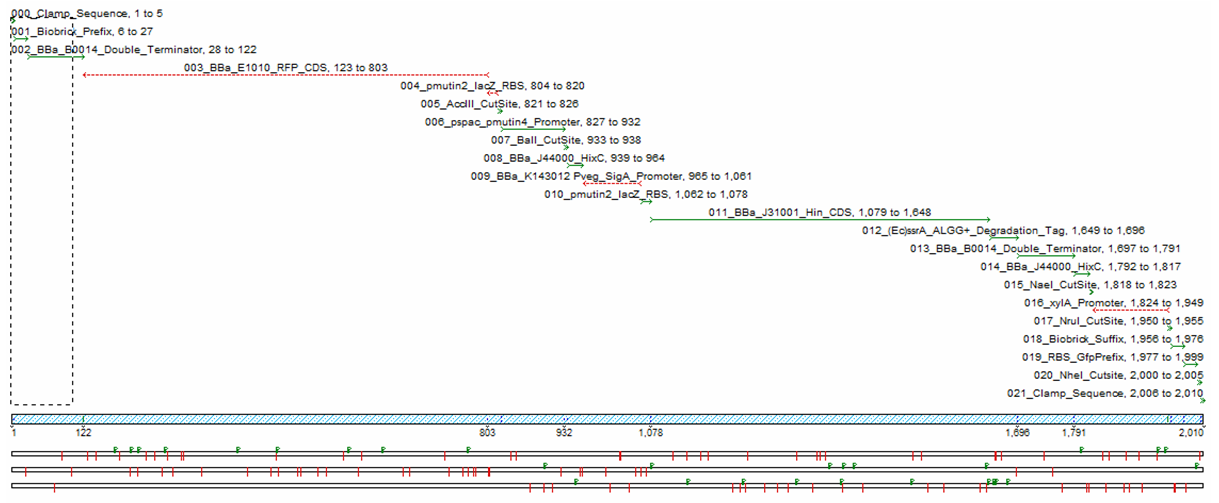
We decided to get our stochastic construct synthesised, as trying to build the construct manually would be too time consuming. The following sequencher diagram shows the components of the construct we had synthesised.
Testing construct
In order to test our construct we had to redesign using inducible promoters governing Hin invertase expression. We used the promoters pSpac and pxylA (Induced by IPTG and Xylose) to test our system. We include cut sites around these promoters in order to replace them with SigmaA promoters once the construct has been characterised.(See sequencher diagram above)
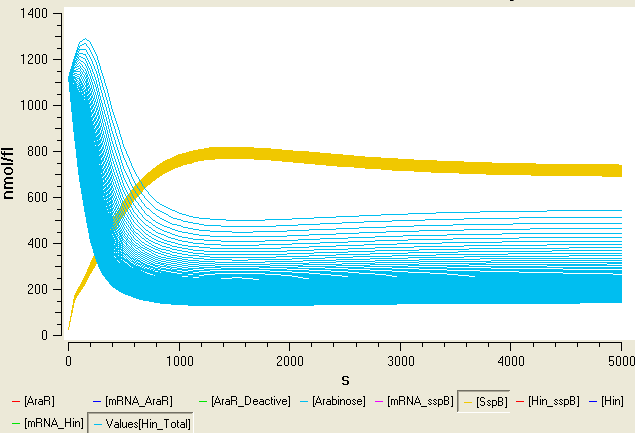
Degradation controller
In order to have another level of control over the orientation of the promoter within the flipping region we added a degradation tag to the Hin invertase protein. The following paper describes how proteins including modified ssrA tags can be located to the ClpXP protease by an Sspb protein. This means that inducible Sspb expression can requlate degradation levels of the tagged protein.
[http://www3.interscience.wiley.com/journal/121415079/abstract?CRETRY=1&SRETRY=0 Inducible protein degradation in Bacillus subtilis using heterologous peptide tags and adaptor proteins to target substrates to the protease ClpXP ]
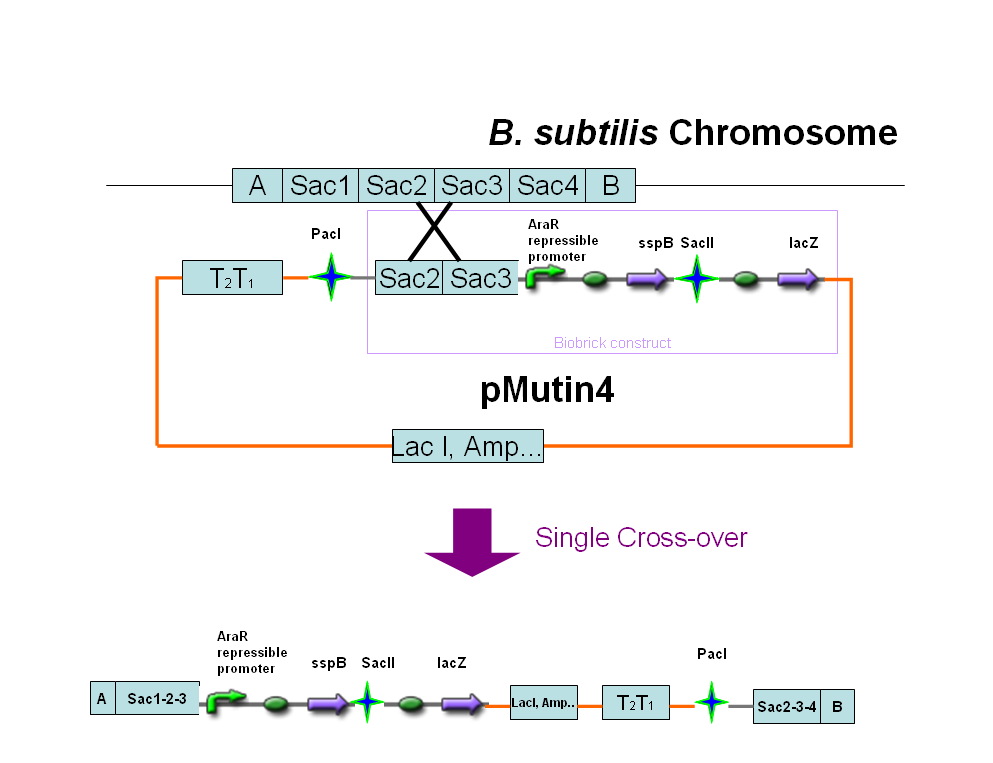
We decided to put the Sspb protein under the control of an arabinose inducible promoter as the following diagram illustrates. We also included a region of the sac gene in our construct, so that the region will integrate into the Bacillus genome at a region other than amyE.
We added a modified version of ssrA degradation tag onto the C-terminus of the Hin protein. Expressed proteins are therefore degraded by ClpXP. However mutations on the ssrA tag weaken the recognition by ClpX, and the modified tags require the SspB adaptor protein to be recognized. When the SspB protein is expressed the proteins tagged with modified version of ssrA tag are targeted for degradation. Otherwise they remain stable.
In B. subtilis there is no sspB orthologue and SspB from E. coli works in B. subtilis. By regulating the levels of SspB by arabinose, we implemented an inducable protein degradation device.
The wild type E. coli ssrA tag is AANDENY-ALAA (SspB recognition site – ClpX recognition site). As suggested in the paper, we took one of the modified ssrA tags to use in our system.
AANDENY-SENY-ALGG (SspB recognition site – SENY +4 Linker - ClpX recognition site)
This tag works well in B. subtilis. However, degradation tags can affect the activity of proteins. Different degradation tags may effect the activity of different proteins. It has been shown that this tag effected the activity of ComA(1).
- Griffith, K. L., and A. D. Grossman. 2008. Inducible protein degradation in Bacillus subtilis using heterologous peptide tags and adaptor proteins to target substrates to the protease ClpXP. Mol. Microbiol. 70:1012-1025.
Stochastic Modelling Tools
Matlab can be used for stochastic modelling. The Glasgow team used Matlab to implement the Gillespie algorithm for incorporating noise among cells. They also used deterministic modelling using ODEs and compared their results. When the number of cells increase two approaches become similar since the noise is cancelled out.
Stocks 2 is another stochastic simulation tool which also uses Gillespie’s direct method and supports SBML.
We used computational modelling in Matlab to try to determine how to make our system tuneable.
Please see our modelling page for Matlab files on our stochastic switch model.
FimE switch
The FimE switch is a similar switch to the Hix system. However, it acts as a latch, meaning that once flipped the segmant will not flip back.
- [http://genomics.lbl.gov/Stuff/TimHam-BandB-online%20version.pdf fimE switch for DNA re-arrangement]
A Tightly Regulated Inducible Expression System Utilising the fim Inversion Recombination Switch.(E. Coli) Timothy S. Ham, Sung Kuk Lee, Jay D. Keasling,Adam P. Arkin,Received 21 December 2005; accepted 2 March 2006 Published online 13 March 2006 in Wiley InterScience (www.interscience.wiley.com). DOI: 10.1002/bit.20916
We decided to use FimE to switch off or on the production of a protein of our choice, such as the genes involved in germination.
- [http://jb.asm.org/cgi/reprint/183/14/4190?maxtoshow=&HITS=10&hits=10&RESULTFORMAT=&fulltext=subtilis&searchid=1&FIRSTINDEX=880&resourcetype=HWFIG Control of the Arabinose Regulon in Bacillus subtilis by AraR In Vivo: Crucial Roles of Operators, Cooperativity, and DNA Looping]
- [http://ukpmc.ac.uk/articlerender.cgi?artid=310841 Binding of the Bacillus subtilis spoIVCA product to the recombination sites of the element interrupting the sigma K-encoding gene] =>...DNA rearrangement that depends on the spoIVCA gene product...
Bistability in Bacillus subtilis
Read this page to find more options for natural stochastic switches in Bacillus subtilis. Natural stochastic switches:Bistability in Bacillus subtilis
And to find out how we are tuning sporulation using our stochastic switch choice see the sporulation tuning page.
Lab strategies
To carry out our labwork we needed cloning strategies for all of our bricks and devices. Please see our cloning strategies page for details on how we cloned our devices.
| | |
|---|---|
| Date: | Achievement: |
| 11/09/09 | Successfully cloned the sspB degradation controller fragment into pSB1AT3 |
| 18/09/09 | Sucessfully cloned the ara promoter/ operator fragment into pSB1AT3 |
| 24/09/09 | Successfully cloned the sspB fragment into the ara + pSB1AT3 prepared backbone. We now have an arabinose inducible degradation controller! |
News
Events
- 20 – 21 June 2009 - Europe workshop (London)
- 23 – 24 June 2009 - UK iGEM meetup (Edinburgh)
- 23 October Practice Presentation (Newcastle)
- 23 October T-shirts are ready
- 27 October Practice Presentation (Sunderland)
- 27 October Poster is ready
- 30 October – 2 November 2009 - Jamboree (Boston)
Social Net
- Newcastle iGEM Twitter
- [http://www.facebook.com/home.php#/group.php?gid=131709337641 Newcastle on Facebook]
- [http://www.youtube.com/user/newcastle2009igem Newcastle Youtube Channel]
 "
"