Team:Purdue/Project
From 2009.igem.org
(→What We've Done So Far) |
|||
| (23 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
[[Image:Header.png]] | [[Image:Header.png]] | ||
| - | <span style="font-size: | + | <span style="font-size:16px; font-family:georgia; border:2px solid black; padding: 6px; margin: 6px; margin-left:15px;">[[Team:Purdue|<span style="color:green;">Home</span>]]</span> |
| - | <span style="font-size: | + | <span style="font-size:16px; font-family:georgia; border:2px solid black; padding: 6px; margin: 6px;">[[Team:Purdue/Team|<span style="color:green;">The Team</span>]]</span> |
| - | <span style="font-size: | + | <span style="font-size:16px; font-family:georgia; border:2px solid black; padding: 6px; margin: 6px;">[[Team:Purdue/Project|The Project]]</span> |
| - | <span style="font-size: | + | <span style="font-size:16px; font-family:georgia; border:2px solid black; padding: 6px; margin: 6px;">[[Team:Purdue/Parts|<span style="color:green;">Parts Submitted to the Registry</span>]]</span> |
| - | <span style="font-size: | + | <span style="font-size:16px; font-family:georgia; border:2px solid black; padding: 6px; margin: 6px;">[[Team:Purdue/Modeling|<span style="color:green;">Modeling</span>]]</span> |
| - | <span style="font-size: | + | <span style="font-size:16px; font-family:georgia; border:2px solid black; padding: 6px; margin: 6px;">[[Team:Purdue/Notebook|<span style="color:green;">Notebook</span>]]</span> |
| + | <span style="font-size:16px; font-family:georgia; border:2px solid black; padding: 6px; margin: 6px;">[[Team:Purdue/Safety|<span style="color:green;">Safety</span>]]</span> | ||
| - | + | == '''Abstract''' == | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | [[Image:PUlogoColor.jpg|450px|center]] | |
| - | ( | + | Glioblastoma multiforme (GBM) is one of the most common forms of primary brain cancer, which usually results in fatality. To date, it has been difficult to overcome primary brain cancer resulting from GBM, primarily because the cancer-initiating cells are suspected to be highly resistant to current cancer therapies. The present study is focused on CD133+ cells found in primary GBM samples. CD133+ cells have shown resistance to hypoxia, irradiation, and some forms of chemotherapy. CD133+ hunting machines will be created by genetically engineering microglial cells (BV-2) with two constructs using mammalian expression vectors. The project will also take advantage of inherent qualities of the microglia such as constant environmental sensing and quick motility. The engineered BV-2s will be equipped to locate the specific GBMs and label the targeted cells with a tat-GFP fusion protein. To create an in-vivo-like environment, the cells will be grown in 3D collagen media. This would create a maze and challenge the microglia to seek out the cancer cells. Cell sorting techniques will be used to measure the accuracy in hitting positive targets, the CD133+ cells. It is the goal of this study to show an alternative approach to cancer treatment, and to emphasize the power of biologically available options to fight the disease. |
| - | == ''' | + | == '''Project Details'''== |
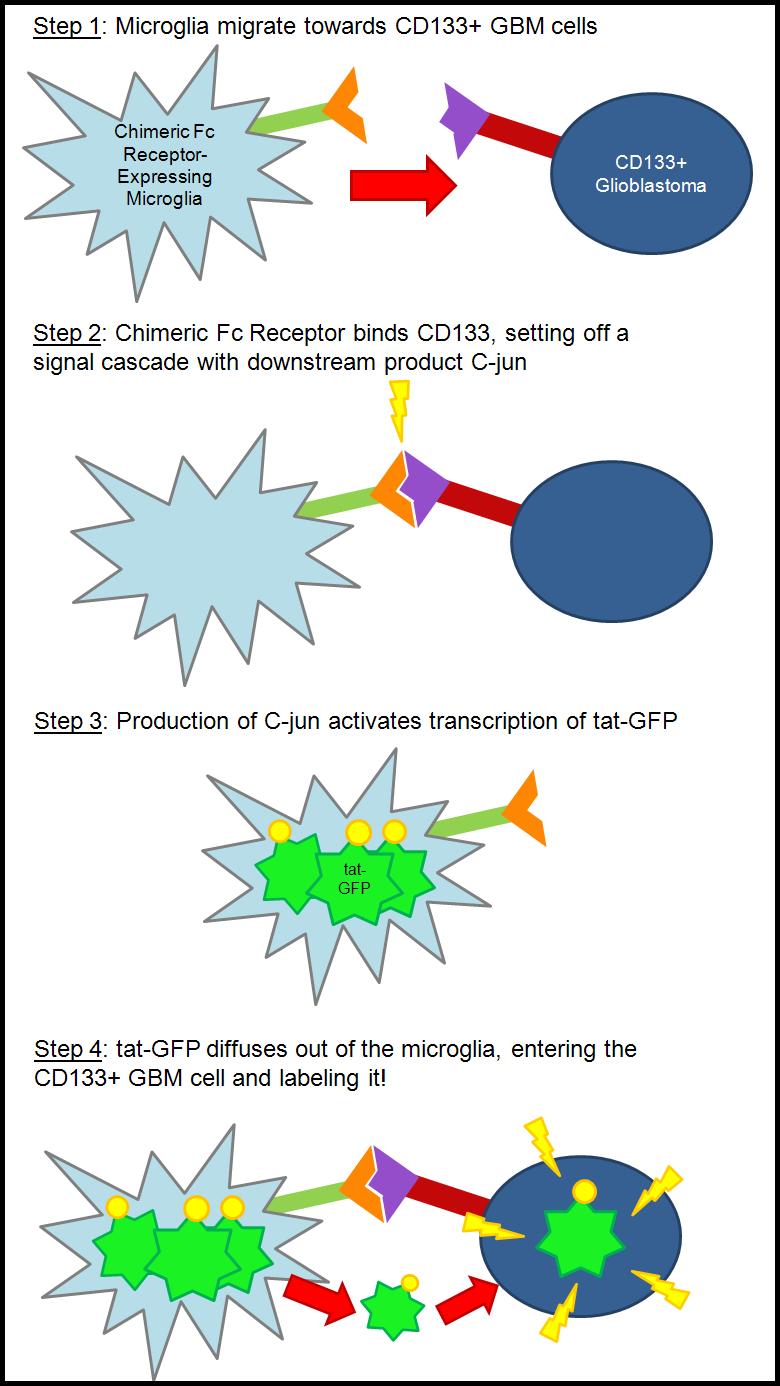
| - | + | === The Concept === | |
| - | + | [[Image:ConceptFlowchart.JPG|400px|left]] | |
| + | Our basic goal is to use the natural mobility of a microglial cell to find specific cells found in 1% of some glioblastoma multiforme. | ||
| + | Glioblastoma multiforme (GBM) is a deadly brain cancer found characterizing over 60% of brain cancers. With current treatment options, even the best-cared for patient, once diagnosed, has little over one year to live. Most recently, Ted Kennedy died of this cancer. | ||
| + | Scientists have found that GBM is made up of many different cell types, some of which make this primary tumor difficult to destroy. In the center of some of these tumor masses, there is a small subset of cells that display stem cell-like qualities. These cells express CD133, a protein found only on stem cells. These stem-like cells have been shown to be resistant to hypoxia, irradiation, and some chemotherapies. These qualities indicate to some a possible reason why recurrence is so high in patients diagnosed with GBM: it is possible that these stem-like cells are the crucial tumor-initiating cells (Cancer Stem Cell Theory), but they are not destroyed along with rest of the tumor mass using current therapies. | ||
| + | Microglial cells are the macrophage of the central nervous system. In a resting state, the microglia have extensions that constantly survey the environment. The microglia interpret signals from the environment and act accordingly. Also, microglia have the ability to "clean up" dead cells and debris via phagocytosis, and move around towards possible threats. | ||
| + | Our goal is to use the microglia to seek out the CD133+ stem-like cells found in the glioblastoma. We plan to achieve this goal by creating a chimeric receptor that is specific for the CD133 protein. Currently, the CD133 protein is not fully characterized. To solve this dilemma, we will use part of the CD133 antibody (Fab region) and most of the Fc gamma receptor to create a very specific chimeric receptor. The Fc receptor is native to the microglia, and thus the microglia have the pathway needed to activate a signal cascade, of which a downstream product is the protein c-jun. As of right now, we are planning on using the c-jun promoter to activate our product, a tat-GFP fusion protein. | ||
| - | + | Tat is a cell-penetrating peptide that was first discovered by two independent labs studying HIV-1. Although the mechanism is unknown, tat has the ability to cross cell membranes with ease. Using this inherent quality, we are attaching it to GFP to create a fusion protein that would label the CD133 cells. The basic idea is that if the CD133+ cell can be labeled, it can be killed. | |
| + | In the future, we would like to hook up an apoptotic factor to the tat in an attempt to kill the CD133+ glioblastoma cells (as opposed to just labeling them). | ||
| + | Overall, we have created a seek and destroy system that is modular and can be adapted to other problems in the human body. | ||
| - | |||
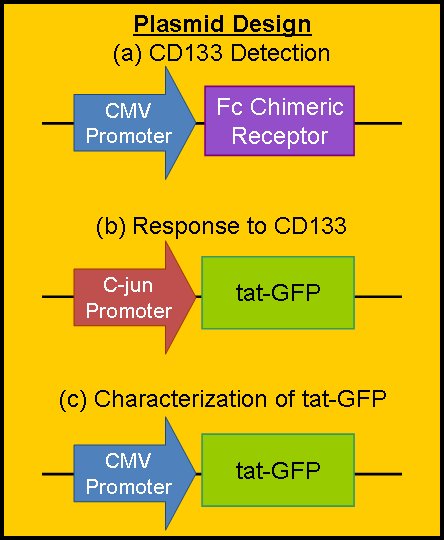
| + | === Plasmid Design === | ||
| + | [[Image:PUPlasmidDesign.jpg|300px|left]] | ||
| + | The Purdue team has design three separate plasmid constructs to accomplish our objectives. All three will of course be inserted into a mammalian plasmid vector with an appropriate antibiotic for selection purposes. | ||
| - | + | Designs (a) and (b) will be used in the final project construct to accomplish our main goal: to seek out and label CD133+ GBM cells. They will hopefully be constructed in the near future. | |
| + | Design (a) encodes for the constitutive production of our chimeric Fc Receptor. The CMV promoter is from a cytomegalovirus, and causes constitutive expression in mammalian cells. The chimeric Fc receptor will be constructed of the Fab portion of the CD133 antibody attached to the body of the Fc gamma receptor found naturally in microglial cells. As such, the receptor will detect the CD133 protein and start a natural signal cascade in the microglia, with a downstream product C-jun. | ||
| + | Design (b) encodes for the production of tat-GFP. The promoter is activated by the protein C-jun, which will be amplified in our system by CD133 detection. As such, the production of tat-GFP will be somewhat leaky (because C-jun will be made by the microglia for other reasons), but the amplification of the gene will be strong when we want it to be so. | ||
| + | Design (c) will be used for the characterization of tat-GFP in our system. By forcing constitutive production of tat-GFP, we can measure kinetic parameters such as rate of production (by using a photobleaching assay), rate of diffusion, and fluorescent intensity vs. concentration of tat-GFP. We have constructed this part for this year's competition. You can view its sequence and details on the [[Team:Purdue/Parts|Purdue Parts Submitted]] page or on the [http://partsregistry.org/Part:BBa_K217000 BBa_K217000 page] in the Registry of Standard Biological Parts. | ||
| - | == | + | === What We've Done So Far === |
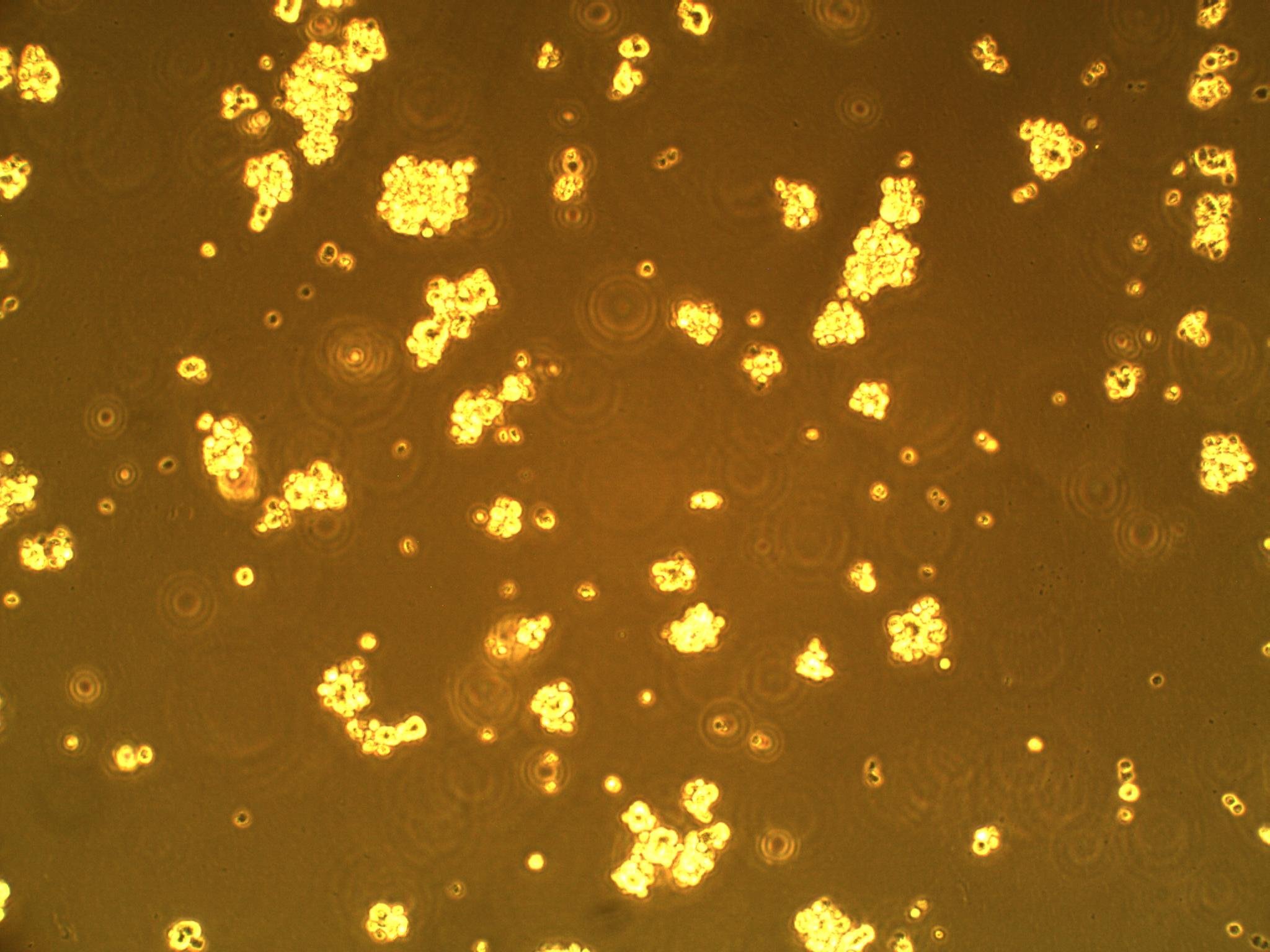
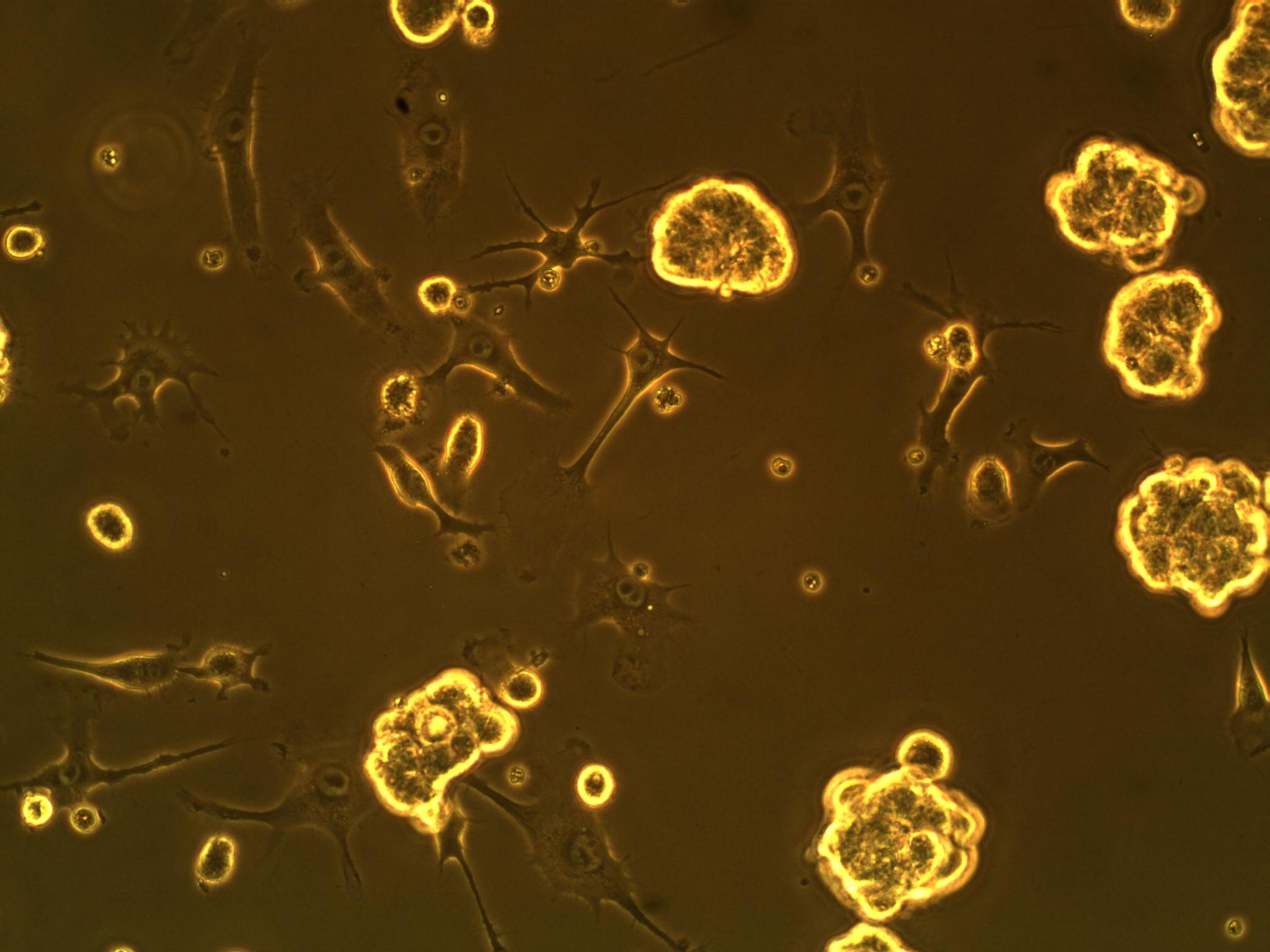
| + | So far, we have cultured our microglial (BV-2) and CD133+ GBM cells (more specifically, GAMB-1 cells derived from human primary brain cancer tissue) in both 2D and 3D to observe any cellular morphological changes that occur between the two types of culture, as they simulate different environments. We've noticed that in 2D culture, the BV-2's attach to the plate, and become flat; in 3D, they are more spherical in geometry. We have also noticed that the CD133+ GBM cells form neurospheres (clusters of cells) in culture--a natural (but interesting) occurrence (see picture below). | ||
| + | {|align = "center" | ||
| + | |- | ||
| + | | | ||
| + | |- | ||
| + | |[[Image:NeurospheresPU.jpg|center|350px|]] ||||||| [[Image:BV2 3D.JPG|center|350px|]] | ||
| + | |- | ||
| + | |CD133+ glioblastoma multiforme cells forming neurospheres ||||||| BV-2 cells in 3D culture (note round morphology) | ||
| + | |- | ||
| + | |} | ||
| - | + | [[Image:GAMB1 3D.JPG|center|350px|]] | |
| - | + | {|align = "center" | |
| - | + | |- | |
| - | + | |GAMB-1 cells in 3D culture (note the different z-range views of neurospheres) | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|} | |} | ||
| + | |||
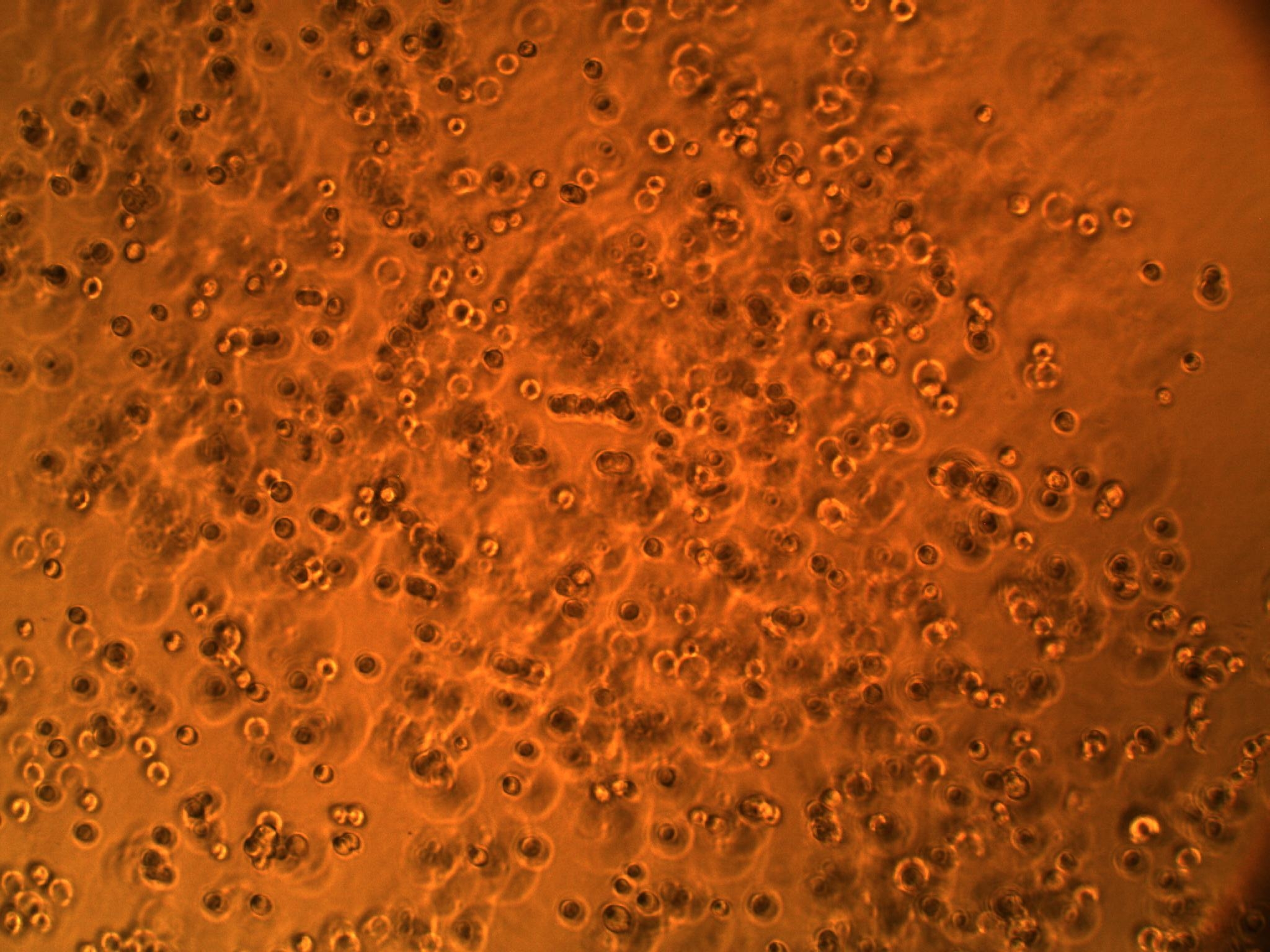
| + | A co-culture of both cell types has also been performed to see how the cells interact in the same setting. We have seen that the BV-2's actually interact with the GBM cells (see picture below). We think they might be attracted to something the GBM cells are secreting (an interleukin, maybe?). | ||
| + | |||
| + | {|align = "center" | ||
| + | |- | ||
| + | | | ||
| + | |- | ||
| + | |[[Image:CoCulture.JPG|center|350px|]] ||||||| [[Image:EcoliPlatePU.jpg|center|350px|]] | ||
| + | |- | ||
| + | |Interaction of BV-2 & GBM cells in 2D co-culture ||||||| Successful transformed colonies of tat-GFP-producing ''E. coli'' | ||
| + | |} | ||
| + | |||
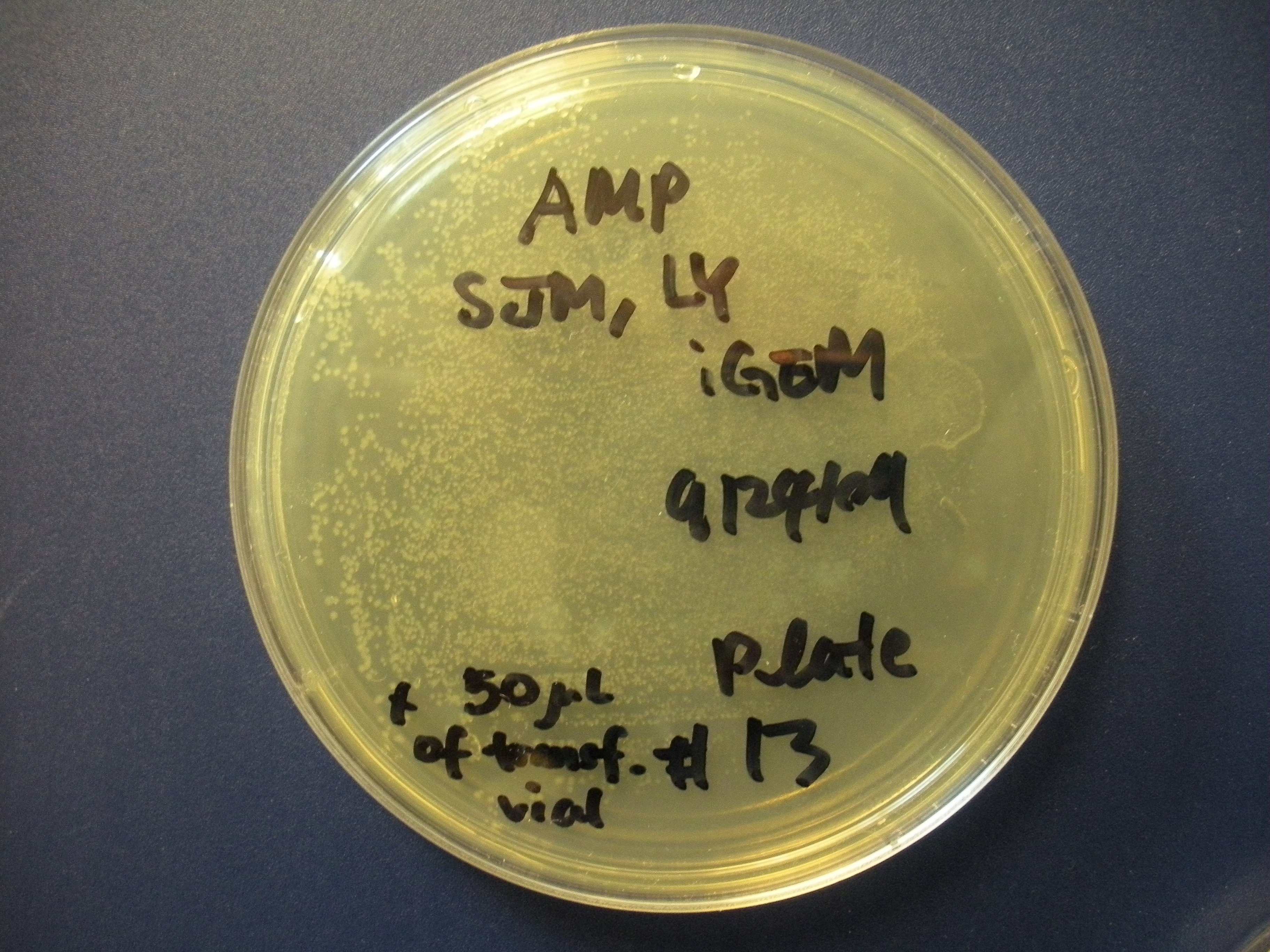
| + | We have also successfully transformed a bacterial version of our new part into ''E. coli'', and we hope to harvest the tat-GFP from them soon for other experiments. In terms of modeling, we have created a working model (see the [[Team:Purdue/Modeling|Modeling Page]]), and we have determined growth rate parameters experimentally for it. | ||
| + | |||
| + | === Future Plans === | ||
| + | |||
| + | The harvested prokaryotic tat-GFP fusion protein will serve as an assay, allowing us to determine the kinetic parameters for our system. As opposed to many other known alternative treatments, it has not escaped our attention that tat-GFP labeled cells show normal morphology (Maddika 2005), a property especially appreciated and crucial for any conclusions to be made in the course of our research. | ||
| + | |||
| + | Based on the experimental data obtained in the previous step, a model will be designed to demonstrate how the dynamics of the system depend on the choice of parameters. By comparing the experimentally obtained data set with the expected theoretical one, the analysis will serve us to either verify the model or encourage its further refinement. A bifurcation analysis will also be performed in order to find crucial parameter values that would drastically change the behavior of the system. | ||
| + | |||
| + | In the subsequent step, the planned plasmids, including the planned chimeric receptors will be created and consequently characterized. | ||
| + | |||
| + | In the final stage, we will then hopefully be able to modify the system accordingly, to serve medical purposes: identification and elimination of CD133+ cells and hence contributing towards preventing the fatal consequences of Glioblastoma multiforme. | ||
| + | |||
| + | |||
| + | === References === | ||
| + | #Alexis, F. et al. (2006). Covalent Attachment of Low Molecular Weight Poly(ethylene imine) Improves Tat Peptide Mediated Gene Delivery. Advanced Materials, 18, 2174-2178. | ||
| + | #Blasi, E. et al. (1989). Immortalization of murine microglial cells by a v-raf/ v-myc carrying retrovirus. Journal of Neuroimmunology, 27, 229-237. | ||
| + | #Holland, E. (2000). Glioblastoma multiforme: The terminator. Proceedings of the National Academy of Science, 97(12), 6242-6244. | ||
| + | #Kim, D. et al. (2009). Generation of Human Induced Pluripotent Stem Cells by Direct Delivery of Reprogramming Proteins. Cell Stem Cell, 4, 472-476. | ||
| + | #Küster, H. et al. (1990). Characterization and Expression of the Gene for the Human Fc Receptor γ Subunit. The Journal of Biological Chemistry, 267(11), 6448-6452. | ||
| + | #Luca, M. (2003). Chemotactic Signaling, Microglia, and Alzheimer’s Disease Senile Plaques: Is There a Connection?. Bulletin of Mathematical Biology, 65, 693-730. | ||
| + | #Martyn, J. et al. Surface display of IgG Fc on baculovirus vectors enhances binding to antigen-presenting cells and cell lines expressing Fc receptors. Archives of Virology, 154, 1129-1138. | ||
| + | #McCord, A. et al. (2009). Physiologic Oxygen Concentration Enhances the Stem-Like Properties of CD133+ Human Glioblastoma Cells In Vitro. Molecular Cancer Research, 7(4), 489-497. | ||
| + | #Park, J. et al. (2007). Adoptive Transfer of Chimeric Antigen Receptor Re-directed Cytolytic T Lymphocyte Clones in Patients with Neuroblastoma. Molecular Therapy, 15(4), 825-833. | ||
| + | #Rock, B. et al. (2004). Role of Microglia in Central Nervous System Infections. Clinical Microbiology Reviews, 17(4), 942-964. | ||
| + | #Qiang, L. et al. (2009). Isolation and characterization of cancer stem like cells in human glioblastoma cell lines. Cancer Letters, 279, 13-21. | ||
| + | #Qiu, W. et al. (1997). Degradation of Amyloid β-Protein by a Metalloprotease Secreted by Microglia and Other Neural and Non-neural Cells. The Journal of Biological Chemistry, 272(10), 6641-6646. | ||
| + | #Song, X. et al. (2004). Fcγ receptor signaling in primary human microglia: differential roles of PI-3K and Ras/ERK MAPK pathways in phagocytosis and chemokine induction. Journal of Leukocyte Biology, 75, 1147-1155. | ||
| + | #Stauss, H. et al. (2007). Monoclonal T-Cell Receptors: New Reagents for Cancer Therapy. www.moleculartherapy.org, 15(10), 1744-1750. | ||
| + | |||
| + | |||
| + | ---- | ||
Latest revision as of 02:52, 22 October 2009
Home The Team The Project Parts Submitted to the Registry Modeling Notebook Safety
Contents |
Abstract
Glioblastoma multiforme (GBM) is one of the most common forms of primary brain cancer, which usually results in fatality. To date, it has been difficult to overcome primary brain cancer resulting from GBM, primarily because the cancer-initiating cells are suspected to be highly resistant to current cancer therapies. The present study is focused on CD133+ cells found in primary GBM samples. CD133+ cells have shown resistance to hypoxia, irradiation, and some forms of chemotherapy. CD133+ hunting machines will be created by genetically engineering microglial cells (BV-2) with two constructs using mammalian expression vectors. The project will also take advantage of inherent qualities of the microglia such as constant environmental sensing and quick motility. The engineered BV-2s will be equipped to locate the specific GBMs and label the targeted cells with a tat-GFP fusion protein. To create an in-vivo-like environment, the cells will be grown in 3D collagen media. This would create a maze and challenge the microglia to seek out the cancer cells. Cell sorting techniques will be used to measure the accuracy in hitting positive targets, the CD133+ cells. It is the goal of this study to show an alternative approach to cancer treatment, and to emphasize the power of biologically available options to fight the disease.
Project Details
The Concept
Our basic goal is to use the natural mobility of a microglial cell to find specific cells found in 1% of some glioblastoma multiforme.
Glioblastoma multiforme (GBM) is a deadly brain cancer found characterizing over 60% of brain cancers. With current treatment options, even the best-cared for patient, once diagnosed, has little over one year to live. Most recently, Ted Kennedy died of this cancer.
Scientists have found that GBM is made up of many different cell types, some of which make this primary tumor difficult to destroy. In the center of some of these tumor masses, there is a small subset of cells that display stem cell-like qualities. These cells express CD133, a protein found only on stem cells. These stem-like cells have been shown to be resistant to hypoxia, irradiation, and some chemotherapies. These qualities indicate to some a possible reason why recurrence is so high in patients diagnosed with GBM: it is possible that these stem-like cells are the crucial tumor-initiating cells (Cancer Stem Cell Theory), but they are not destroyed along with rest of the tumor mass using current therapies.
Microglial cells are the macrophage of the central nervous system. In a resting state, the microglia have extensions that constantly survey the environment. The microglia interpret signals from the environment and act accordingly. Also, microglia have the ability to "clean up" dead cells and debris via phagocytosis, and move around towards possible threats.
Our goal is to use the microglia to seek out the CD133+ stem-like cells found in the glioblastoma. We plan to achieve this goal by creating a chimeric receptor that is specific for the CD133 protein. Currently, the CD133 protein is not fully characterized. To solve this dilemma, we will use part of the CD133 antibody (Fab region) and most of the Fc gamma receptor to create a very specific chimeric receptor. The Fc receptor is native to the microglia, and thus the microglia have the pathway needed to activate a signal cascade, of which a downstream product is the protein c-jun. As of right now, we are planning on using the c-jun promoter to activate our product, a tat-GFP fusion protein.
Tat is a cell-penetrating peptide that was first discovered by two independent labs studying HIV-1. Although the mechanism is unknown, tat has the ability to cross cell membranes with ease. Using this inherent quality, we are attaching it to GFP to create a fusion protein that would label the CD133 cells. The basic idea is that if the CD133+ cell can be labeled, it can be killed.
In the future, we would like to hook up an apoptotic factor to the tat in an attempt to kill the CD133+ glioblastoma cells (as opposed to just labeling them).
Overall, we have created a seek and destroy system that is modular and can be adapted to other problems in the human body.
Plasmid Design
The Purdue team has design three separate plasmid constructs to accomplish our objectives. All three will of course be inserted into a mammalian plasmid vector with an appropriate antibiotic for selection purposes.
Designs (a) and (b) will be used in the final project construct to accomplish our main goal: to seek out and label CD133+ GBM cells. They will hopefully be constructed in the near future.
Design (a) encodes for the constitutive production of our chimeric Fc Receptor. The CMV promoter is from a cytomegalovirus, and causes constitutive expression in mammalian cells. The chimeric Fc receptor will be constructed of the Fab portion of the CD133 antibody attached to the body of the Fc gamma receptor found naturally in microglial cells. As such, the receptor will detect the CD133 protein and start a natural signal cascade in the microglia, with a downstream product C-jun.
Design (b) encodes for the production of tat-GFP. The promoter is activated by the protein C-jun, which will be amplified in our system by CD133 detection. As such, the production of tat-GFP will be somewhat leaky (because C-jun will be made by the microglia for other reasons), but the amplification of the gene will be strong when we want it to be so.
Design (c) will be used for the characterization of tat-GFP in our system. By forcing constitutive production of tat-GFP, we can measure kinetic parameters such as rate of production (by using a photobleaching assay), rate of diffusion, and fluorescent intensity vs. concentration of tat-GFP. We have constructed this part for this year's competition. You can view its sequence and details on the Purdue Parts Submitted page or on the [http://partsregistry.org/Part:BBa_K217000 BBa_K217000 page] in the Registry of Standard Biological Parts.
What We've Done So Far
So far, we have cultured our microglial (BV-2) and CD133+ GBM cells (more specifically, GAMB-1 cells derived from human primary brain cancer tissue) in both 2D and 3D to observe any cellular morphological changes that occur between the two types of culture, as they simulate different environments. We've noticed that in 2D culture, the BV-2's attach to the plate, and become flat; in 3D, they are more spherical in geometry. We have also noticed that the CD133+ GBM cells form neurospheres (clusters of cells) in culture--a natural (but interesting) occurrence (see picture below).
| CD133+ glioblastoma multiforme cells forming neurospheres | BV-2 cells in 3D culture (note round morphology) |
| GAMB-1 cells in 3D culture (note the different z-range views of neurospheres) |
A co-culture of both cell types has also been performed to see how the cells interact in the same setting. We have seen that the BV-2's actually interact with the GBM cells (see picture below). We think they might be attracted to something the GBM cells are secreting (an interleukin, maybe?).
| Interaction of BV-2 & GBM cells in 2D co-culture | Successful transformed colonies of tat-GFP-producing E. coli |
We have also successfully transformed a bacterial version of our new part into E. coli, and we hope to harvest the tat-GFP from them soon for other experiments. In terms of modeling, we have created a working model (see the Modeling Page), and we have determined growth rate parameters experimentally for it.
Future Plans
The harvested prokaryotic tat-GFP fusion protein will serve as an assay, allowing us to determine the kinetic parameters for our system. As opposed to many other known alternative treatments, it has not escaped our attention that tat-GFP labeled cells show normal morphology (Maddika 2005), a property especially appreciated and crucial for any conclusions to be made in the course of our research.
Based on the experimental data obtained in the previous step, a model will be designed to demonstrate how the dynamics of the system depend on the choice of parameters. By comparing the experimentally obtained data set with the expected theoretical one, the analysis will serve us to either verify the model or encourage its further refinement. A bifurcation analysis will also be performed in order to find crucial parameter values that would drastically change the behavior of the system.
In the subsequent step, the planned plasmids, including the planned chimeric receptors will be created and consequently characterized.
In the final stage, we will then hopefully be able to modify the system accordingly, to serve medical purposes: identification and elimination of CD133+ cells and hence contributing towards preventing the fatal consequences of Glioblastoma multiforme.
References
- Alexis, F. et al. (2006). Covalent Attachment of Low Molecular Weight Poly(ethylene imine) Improves Tat Peptide Mediated Gene Delivery. Advanced Materials, 18, 2174-2178.
- Blasi, E. et al. (1989). Immortalization of murine microglial cells by a v-raf/ v-myc carrying retrovirus. Journal of Neuroimmunology, 27, 229-237.
- Holland, E. (2000). Glioblastoma multiforme: The terminator. Proceedings of the National Academy of Science, 97(12), 6242-6244.
- Kim, D. et al. (2009). Generation of Human Induced Pluripotent Stem Cells by Direct Delivery of Reprogramming Proteins. Cell Stem Cell, 4, 472-476.
- Küster, H. et al. (1990). Characterization and Expression of the Gene for the Human Fc Receptor γ Subunit. The Journal of Biological Chemistry, 267(11), 6448-6452.
- Luca, M. (2003). Chemotactic Signaling, Microglia, and Alzheimer’s Disease Senile Plaques: Is There a Connection?. Bulletin of Mathematical Biology, 65, 693-730.
- Martyn, J. et al. Surface display of IgG Fc on baculovirus vectors enhances binding to antigen-presenting cells and cell lines expressing Fc receptors. Archives of Virology, 154, 1129-1138.
- McCord, A. et al. (2009). Physiologic Oxygen Concentration Enhances the Stem-Like Properties of CD133+ Human Glioblastoma Cells In Vitro. Molecular Cancer Research, 7(4), 489-497.
- Park, J. et al. (2007). Adoptive Transfer of Chimeric Antigen Receptor Re-directed Cytolytic T Lymphocyte Clones in Patients with Neuroblastoma. Molecular Therapy, 15(4), 825-833.
- Rock, B. et al. (2004). Role of Microglia in Central Nervous System Infections. Clinical Microbiology Reviews, 17(4), 942-964.
- Qiang, L. et al. (2009). Isolation and characterization of cancer stem like cells in human glioblastoma cell lines. Cancer Letters, 279, 13-21.
- Qiu, W. et al. (1997). Degradation of Amyloid β-Protein by a Metalloprotease Secreted by Microglia and Other Neural and Non-neural Cells. The Journal of Biological Chemistry, 272(10), 6641-6646.
- Song, X. et al. (2004). Fcγ receptor signaling in primary human microglia: differential roles of PI-3K and Ras/ERK MAPK pathways in phagocytosis and chemokine induction. Journal of Leukocyte Biology, 75, 1147-1155.
- Stauss, H. et al. (2007). Monoclonal T-Cell Receptors: New Reagents for Cancer Therapy. www.moleculartherapy.org, 15(10), 1744-1750.
 "
"