Team:Sheffield/Modeling
From 2009.igem.org
Kirubhakaran (Talk | contribs) |
Kirubhakaran (Talk | contribs) |
||
| Line 116: | Line 116: | ||
===Wet lab result analysis=== | ===Wet lab result analysis=== | ||
| + | [[Image:result1.png|center|900px|border]] | ||
| - | + | From the graph above we could see that the system does behave as our predicted model after 12 hours. The system tends to be unstable before it. A kinetic model can be defined for the system after 12 hours, which is: | |
| + | For all 3 antibiotics | ||
| + | y= -0.1518x^2 + 1.5289x + 7.5641 | ||
| + | |||
| + | For control | ||
| + | y= -1.5133x^2 + 16.8821x - 11.1247 | ||
| + | |||
| + | Where y is activity of LacZ (miller units) and x is intensity (microeinstein/square meter/ second) | ||
| Line 128: | Line 136: | ||
[[Image:result2.png|center|900px|border]] | [[Image:result2.png|center|900px|border]] | ||
| + | |||
| + | |||
| + | These graphs show a clear idea of how the whole system binds together, which has 3 parameters varying (intensity, time and activity of LacZ). Any design process which involves this system could use this to predict the outcome of the model. | ||
| + | |||
| + | For instance if we are working with this system under a wavelength of 6 (all 3 antibiotics), the time series model would be; | ||
| + | |||
| + | |||
| + | y= -0.4152t2 + 7.6872t -16.4965 | ||
| + | |||
| + | Where y is activity of LacZ (miller units) and t is time (hours) | ||
Revision as of 18:32, 29 September 2009
| Home | Team | Project | Parts | Modeling | Notebook |
|---|
|
SYSTEM BREAKDOWNWe are going to analyse the biological system in a control perspective way. Our 3 main aims are; 1) Suggesting a realistic model for the system 2) Comparing the wet lab result with our model 3) A standard description of how our overall system intends to work.
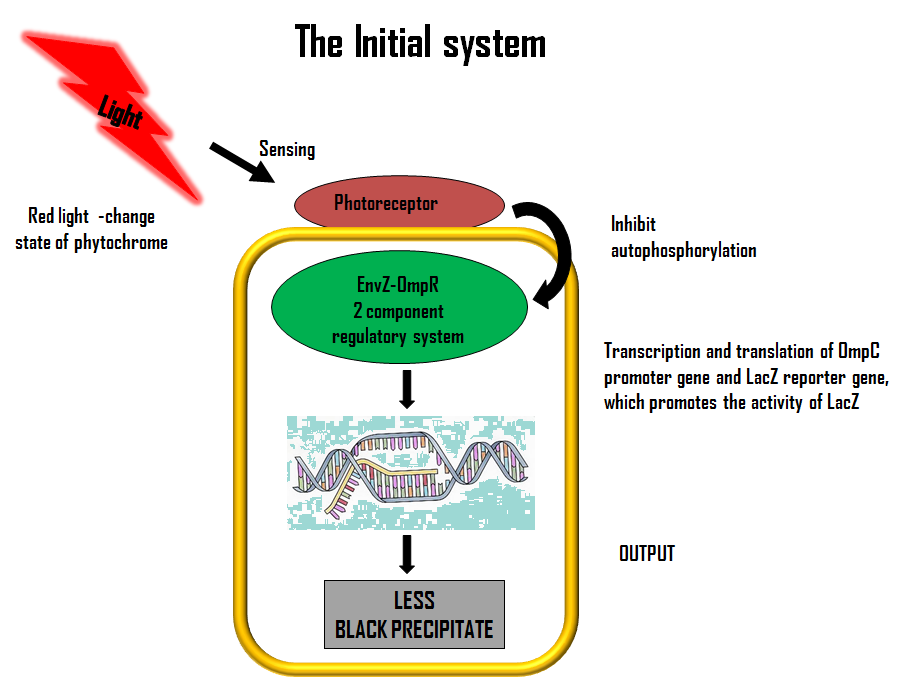
ModelLets first look at how our initial system works. The flow diagram below gives a brief description of it;
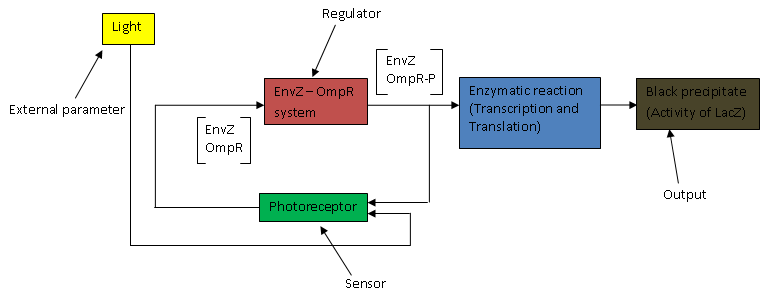
The above flow chart can be reconstructed into a control block diagram;
The script below shows how each block parameters are designed
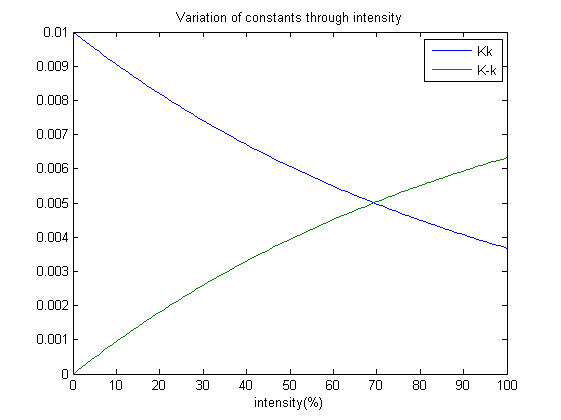
Our next step is to simulate the model, which brings about an interesting design practise which my team has employed. Which is, how does the photoreceptor works? And how does it affect the system when light intensity varies? A point to note; each ODE’s tends to a steady state after some time. Therefore we have suggested that: Each constant varies with the amount of light shined on the system and as well achieves a steady state at a certain point. Without the presence of red light, the concentration of EnvZP, (EnvZP)OmpR and OmpRP should increase. Therefore the constants which affect the rate of reaction of those should decrease with intensity; which is K1, K-2, Kk and Kt. Whereas K-1, K2, K-k and Kp increase as intensity increases.
Initial concentration of: EnvZ = 1M, EnvZP = 1M, (EnvZP)OmpR = 1M, OmpRP = 1M, OmpR = 1M, EnvZ(OmpRP) = 1M.
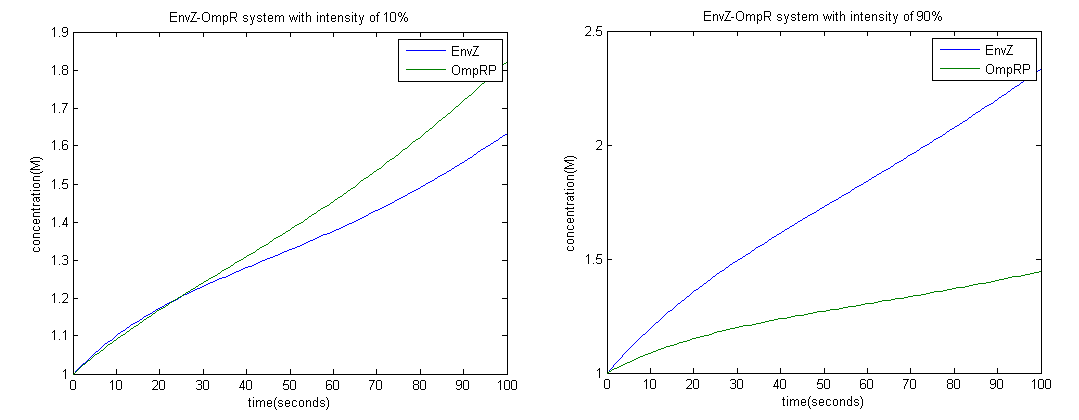
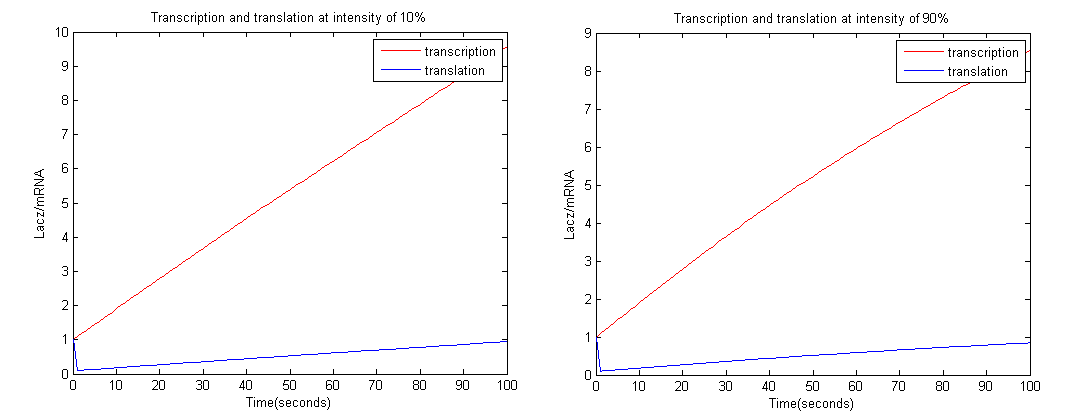
To achieve this we employed the Euler method; with a time interval of 1 second.
From the graph above it is clear that at high intensity OmpRP concentration is less, therefore activity of LacZ should be less as well.
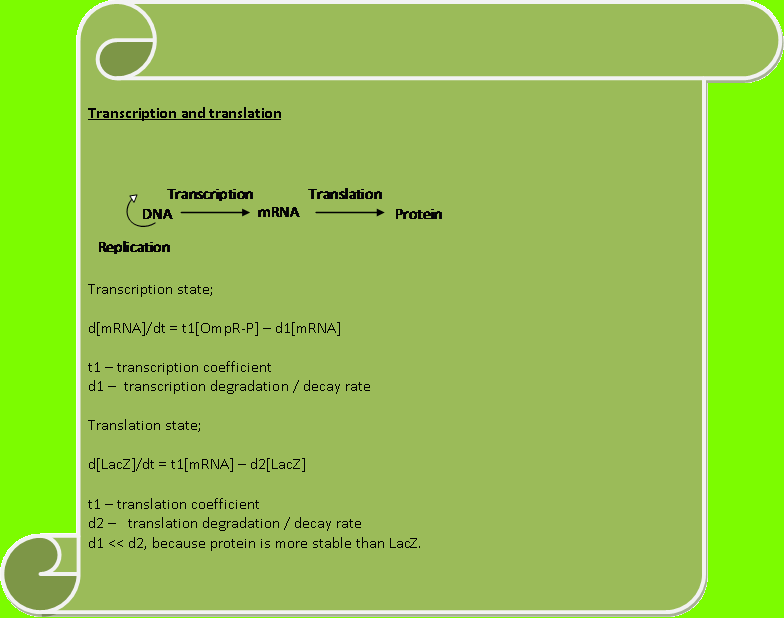
Constants used: t1=0.1, t2=0.1, d1=0.01, d2=1.
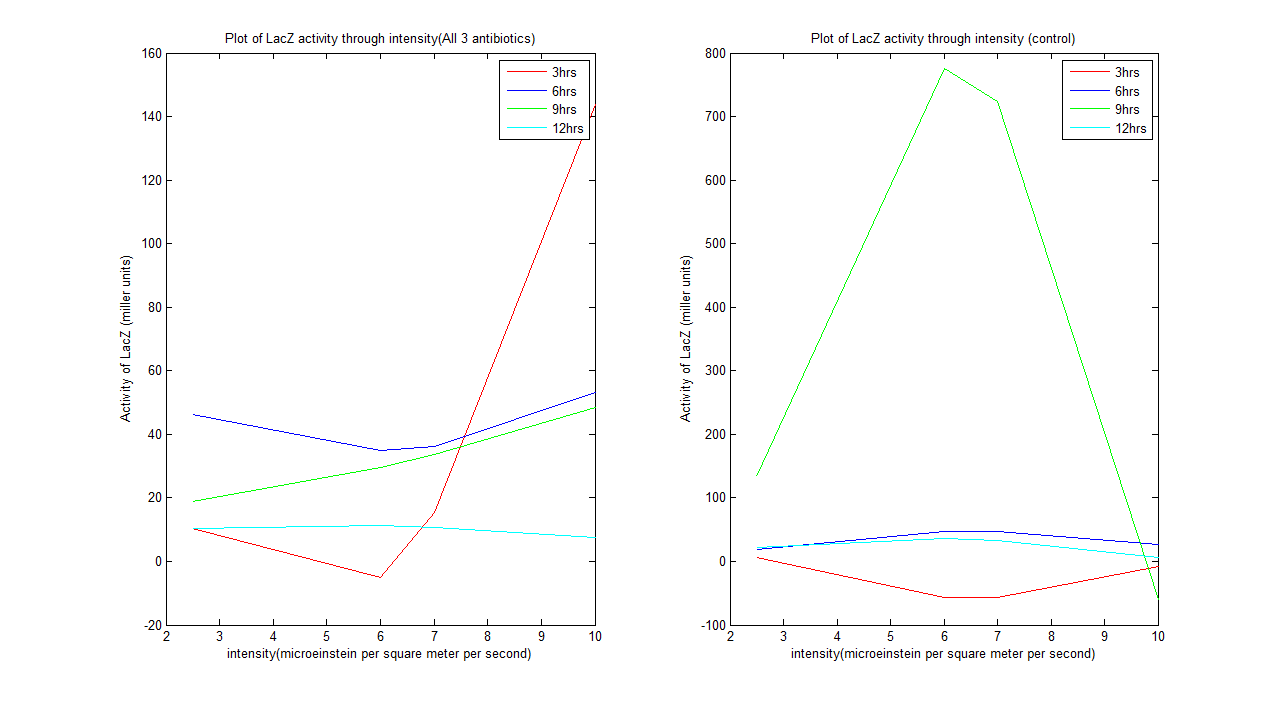
Wet lab result analysis
From the graph above we could see that the system does behave as our predicted model after 12 hours. The system tends to be unstable before it. A kinetic model can be defined for the system after 12 hours, which is: For all 3 antibiotics y= -0.1518x^2 + 1.5289x + 7.5641 For control y= -1.5133x^2 + 16.8821x - 11.1247 Where y is activity of LacZ (miller units) and x is intensity (microeinstein/square meter/ second)
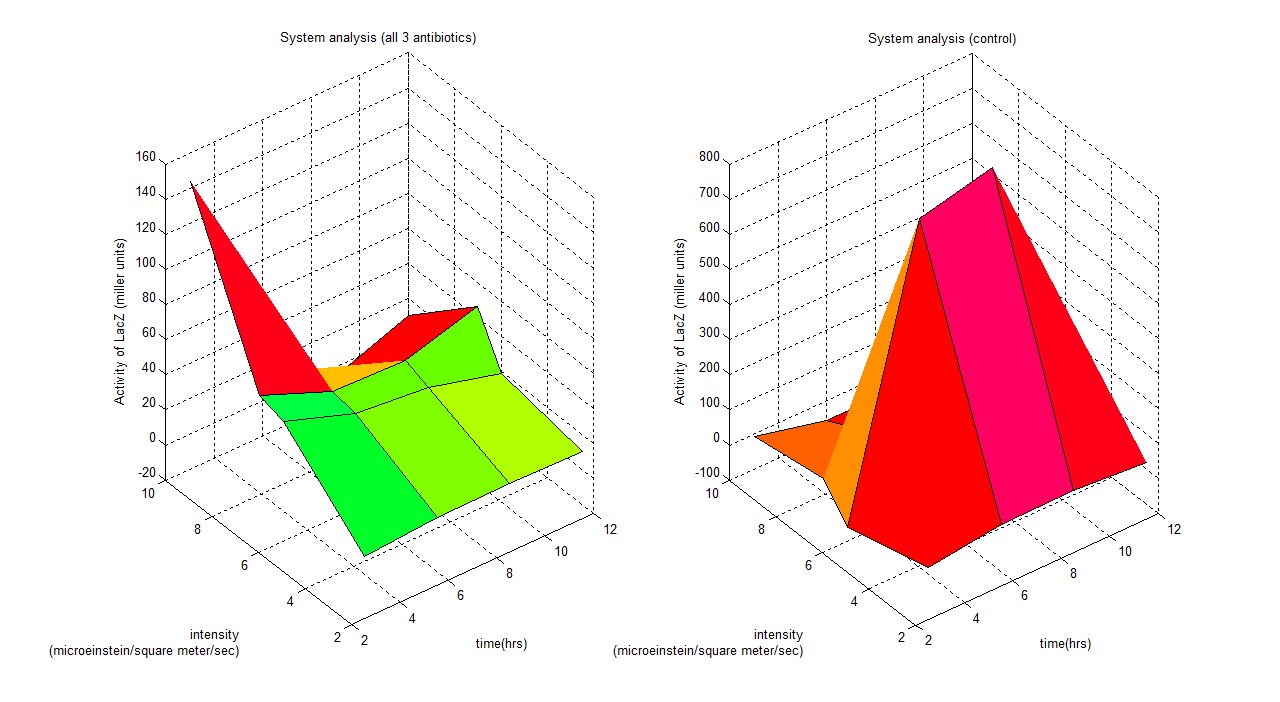
These graphs show a clear idea of how the whole system binds together, which has 3 parameters varying (intensity, time and activity of LacZ). Any design process which involves this system could use this to predict the outcome of the model. For instance if we are working with this system under a wavelength of 6 (all 3 antibiotics), the time series model would be;
Where y is activity of LacZ (miller units) and t is time (hours)
General descriptionReferences[http://www.pnas.org/content/100/2/691.full# 1) Robustness and the cycle of phosphorylation and dephosphorylation in a two-component regulatory system(Eric Batchelor and Mark Goulian)]
|
 "
"