Team:Sheffield/Project
From 2009.igem.org
(→Project Details) |
(→Overall project) |
||
| (18 intermediate revisions not shown) | |||
| Line 8: | Line 8: | ||
!align="center"|[[Team:Sheffield/Team|Team]] | !align="center"|[[Team:Sheffield/Team|Team]] | ||
!align="center"|[[Team:Sheffield/Project|Project]] | !align="center"|[[Team:Sheffield/Project|Project]] | ||
| - | !align="center"|[[Team:Sheffield/ | + | !align="center"|[[Team:Sheffield/Further Work|Further Work]] |
!align="center"|[[Team:Sheffield/Modeling|Modeling]] | !align="center"|[[Team:Sheffield/Modeling|Modeling]] | ||
!align="center"|[[Team:Sheffield/Notebook|Notebook]] | !align="center"|[[Team:Sheffield/Notebook|Notebook]] | ||
| Line 20: | Line 20: | ||
== '''Overall project''' == | == '''Overall project''' == | ||
| - | In this project, we originally intended to create an E.coli system that is sensitive to multiple wavelengths of light and produce a colour indication of the specific wavelength it is exposed to. However due to limitations in the time and number of people available, and due to unexpected results the project changed significantly. | + | '''The Initial System''' |
| + | |||
| + | |||
| + | [[Image:Sheff_PCB1.jpg|900px|center|boarder]] | ||
| + | |||
| + | |||
| + | In this project, we originally intended to create an E.coli system that is sensitive to multiple wavelengths of light and produce a colour indication of the specific wavelength it is exposed to. We use the initial system that was mentioned inthe Levskaya et al. which involves phycocyanobilin produced through ho1 and pcyA biosynthesis gene converting haem. Phycocyanobilin then forms part of the photoreceptor which controls phosphorylation of the whole system. | ||
| + | |||
| + | |||
| + | However due to limitations in the time and number of people available, and due to unexpected results the project changed significantly. | ||
| Line 55: | Line 64: | ||
[[Image:sheff_results_colourfilter.jpg|center|border]]Graduation of colouration can be seen clearly in this results table, the sample under red filter has the least colouration, while the sample in the dark has the most colouration. Blue and green has similar level of colouration judging by eye. This result matches the theory in the paper; that red light inhibits the gene expression the most. However, to complete the charaterisation of the system, quantitative measurements is required. Miller Assay quantifies the amount of beta-galactocidase, this can act as an indicator of how active the gene expression is. | [[Image:sheff_results_colourfilter.jpg|center|border]]Graduation of colouration can be seen clearly in this results table, the sample under red filter has the least colouration, while the sample in the dark has the most colouration. Blue and green has similar level of colouration judging by eye. This result matches the theory in the paper; that red light inhibits the gene expression the most. However, to complete the charaterisation of the system, quantitative measurements is required. Miller Assay quantifies the amount of beta-galactocidase, this can act as an indicator of how active the gene expression is. | ||
| - | |||
| - | + | '''Varying light intensity''' | |
| - | and then to | + | After verifying that red light inhibits the most gene expression, the characterisation of the intensity can be proceed. According to the research paper, the light intensity can effect the amount of gene expression in this system. To investigate this, 3 sample and a control strain were put into LB broth, was then exposed to red light for 12hrs at the following intensity uEins/m2/s: 2.5, 6, 7, 10. Miller Assay was then performed to measure the amount of beta-galactocidase produced at each intensity by measuring the optical density of each sample in a photo-spectrometer. |
| - | + | ||
| - | + | ||
== Results == | == Results == | ||
| + | '''Characterisation of the system''' | ||
| + | |||
| + | |||
| + | [[Image:result1.png|center|900px|border]] | ||
| + | |||
| + | |||
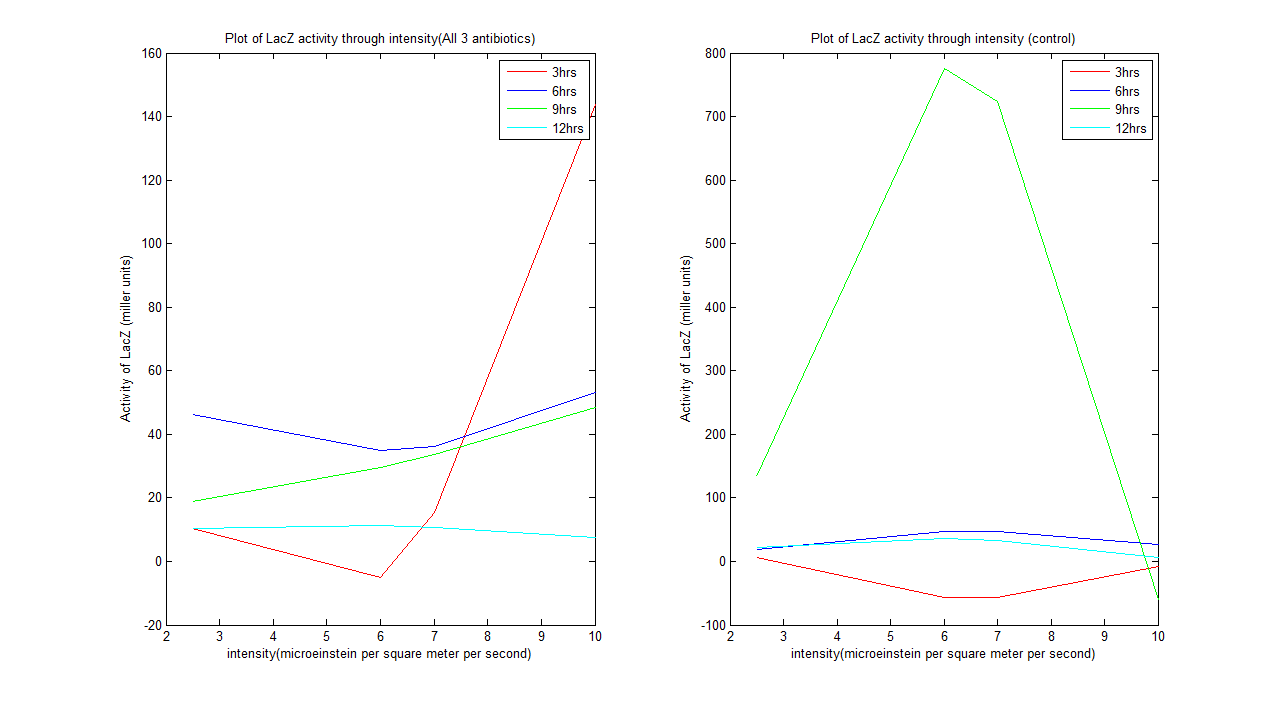
| + | Graphs above shows the model of LacZ activity over different intensity over the course of 12 hours. | ||
| + | |||
| + | - This model is based on our experimental result and it matches with the model in the paper. | ||
| + | |||
| + | - An important point is that the amount of black precipitate the system produce is not a representation of the amount of LacZ activity, because the black precipitate does not degrades on the agar plate. | ||
| + | |||
| + | -Hence why Miller Assay must be carried out to quantify the amount of LacZ activity. | ||
| + | |||
| + | '''Graph analysis''' | ||
| + | |||
| + | '''3hrs'''- The system is very unstable, the beginning of translation and transcription of gene, LacZ activity is erratic, does not form a trend proportional to intensity over the first 3 hours. | ||
| + | |||
| + | '''6hrs'''- The system is still unstable, but relatively calmer. | ||
| + | |||
| + | '''9hrs'''- The system is more similar to expected trend. | ||
| + | |||
| + | '''12hrs'''- The system behaves as expected in the paper, LacZ activity decrease as light intensity increase | ||
| + | |||
| + | The control strain has produced a similar trend to the experimental values. | ||
| + | |||
| + | |||
| + | |||
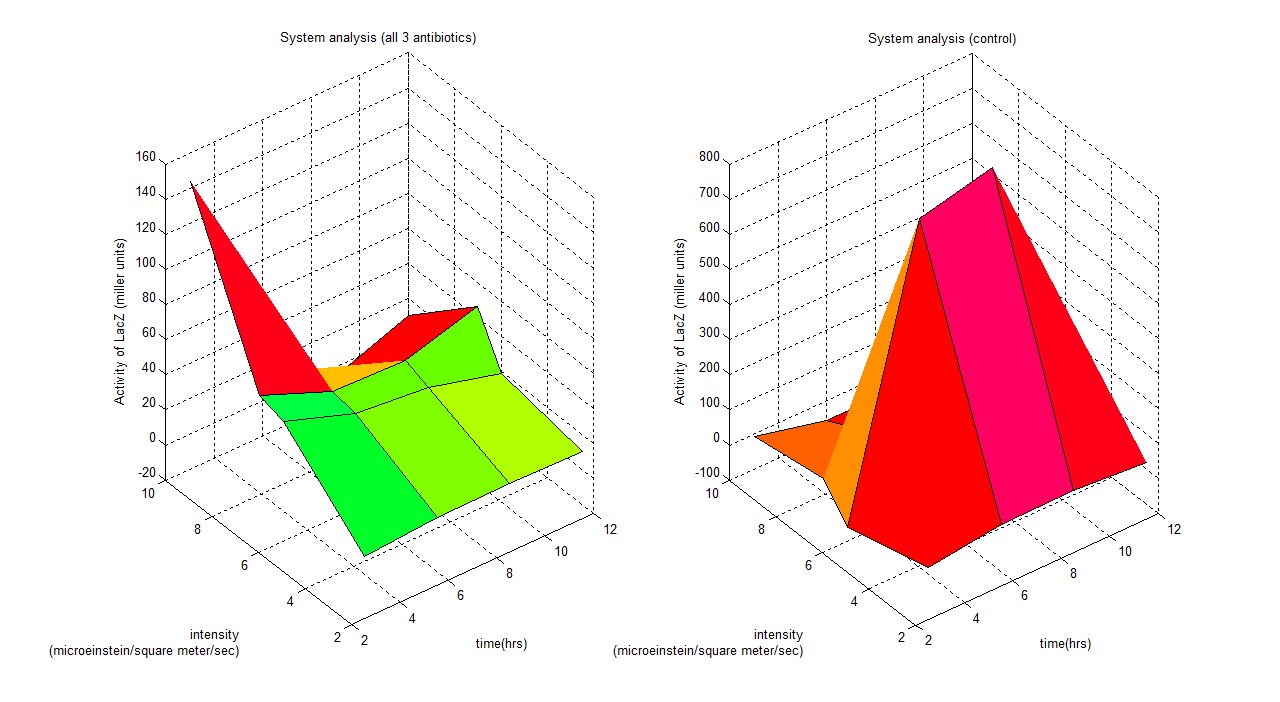
| + | '''3D Characterisation plot''' | ||
| + | |||
| + | [[Image:result2.png|center|900px|border]] | ||
| + | |||
| + | |||
| + | |||
| + | This 3D plot is generated through Matlab, it models the system's LacZ activity over various light intensity over a course of 12 hours. | ||
==Discussion== | ==Discussion== | ||
| - | + | This [[link]] shows the table of measurement taken in order to obtain results shown above. | |
| + | |||
| + | All the results we have got show trends and strong characterization, which gives an idea how to go about the design. | ||
| + | |||
| + | The error bias of our result cannot be calculated or shown but; to obtain a more accurate result more readings have to be taken. | ||
| - | |||
| - | |||
==References== | ==References== | ||
===Papers=== | ===Papers=== | ||
| - | #'''Engineering Escherichia coli to see light'''<br> ''Nature'' 24 November 2005 DOI:10.1038/nature04405<br> A. Levskaya ''et al.''<br> [http://www.nature.com/nature/journal/v438/n7067/full/nature04405.html URL] [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=16306980 Pubmed] [http://www.hubmed.org/display.cgi?uids=16306980 Hubmed]|} | + | #'''Engineering Escherichia coli to see light'''<br> ''Nature'' 24 November 2005 DOI:10.1038/nature04405<br> A. Levskaya ''et al.''<br> [http://www.nature.com/nature/journal/v438/n7067/full/nature04405.html URL] [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=16306980 Pubmed] [http://www.hubmed.org/display.cgi?uids=16306980 Hubmed] |
| + | |||
| + | |} | ||
Latest revision as of 15:04, 19 October 2009
| Home | Team | Project | Further Work | Modeling | Notebook |
|---|
|
Overall projectThe Initial System
Project DetailsThe first experiment we carried to familiarise ourselves with the strain, was to repeat the light sensing ability of RU1012 as described in the Levskaya et al. paper[1], in its most simple form. Red light switch off the photoreceptor through inhibiting autophosphorylation, and therefore switching off the expression for Lac Z production. We shown borad spectrum light on one sample and kept the other in the dark so that gene expression can not be interefered. The samples were put on seperate agar plates with X-gal on it which acts as a substrate for Lac Z and producing a blue colouration. To our surprise, the result were opposite to what we expected: the illuminated plate produced more pigment than the one in the dark, whereas it should have been the opposite. No control was used in this experiment. The same experiment was repeated with a control strain to confirm that the result was systematic. Later on we realise that the wavelength that inhibits the autophosphorylation has to be a specific wavelength, a broadspectrun light that contains that wavelength does not have the same effect. Too solve that problem we have tried various measures:
The Balmer lamp provided a narrower band of wavelength, we hoped that it can characterise the E.coli system better than broad spectrum light. Experiments were repeated with Hydrogen lamp, conducted at room temperature - due to set up of the lamp, the experiment can not be carried out in an incubator at 37C. Unfortunately, this has not changed the outcome of the result, the gene expression was still not switched off.
Filter paper
ResultsCharacterisation of the system
- This model is based on our experimental result and it matches with the model in the paper. - An important point is that the amount of black precipitate the system produce is not a representation of the amount of LacZ activity, because the black precipitate does not degrades on the agar plate. -Hence why Miller Assay must be carried out to quantify the amount of LacZ activity. Graph analysis 3hrs- The system is very unstable, the beginning of translation and transcription of gene, LacZ activity is erratic, does not form a trend proportional to intensity over the first 3 hours. 6hrs- The system is still unstable, but relatively calmer. 9hrs- The system is more similar to expected trend. 12hrs- The system behaves as expected in the paper, LacZ activity decrease as light intensity increase The control strain has produced a similar trend to the experimental values.
3D Characterisation plot
This 3D plot is generated through Matlab, it models the system's LacZ activity over various light intensity over a course of 12 hours. DiscussionThis link shows the table of measurement taken in order to obtain results shown above. All the results we have got show trends and strong characterization, which gives an idea how to go about the design. The error bias of our result cannot be calculated or shown but; to obtain a more accurate result more readings have to be taken.
ReferencesPapers
|
 "
"