Team:TUDelft/Modeling
From 2009.igem.org
(→Mathematical Modeling) |
(→Delay Device) |
||
| Line 3: | Line 3: | ||
='''Delay Device'''= | ='''Delay Device'''= | ||
[[Image:Negative Feedforward.jpg]]<br> | [[Image:Negative Feedforward.jpg]]<br> | ||
| - | Figure 1: Cascade model<br> | + | Figure 1: Negative Cascade model<br> |
[[Image:Riboregulator_delay.jpg]]<br> | [[Image:Riboregulator_delay.jpg]]<br> | ||
Figure 2: Riboregulator model<br> | Figure 2: Riboregulator model<br> | ||
==Mathematical Modeling== | ==Mathematical Modeling== | ||
| - | The Cascade model shown above can be expressed with the following set of ODEs (ordinary differential equations):<br> | + | The Negative Cascade model shown above can be expressed with the following set of ODEs (ordinary differential equations):<br> |
[[Image:negative_feedforward_ODEs_2.jpg]]<br><br> | [[Image:negative_feedforward_ODEs_2.jpg]]<br><br> | ||
The Riboregulator model shown above can be expressed with the following set of ODEs:<br> | The Riboregulator model shown above can be expressed with the following set of ODEs:<br> | ||
Revision as of 12:57, 7 August 2009
Delay Device
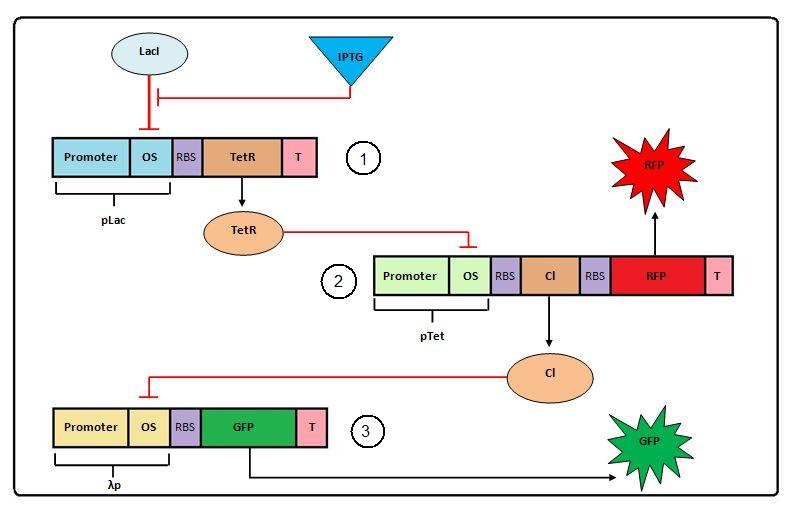
Figure 1: Negative Cascade model
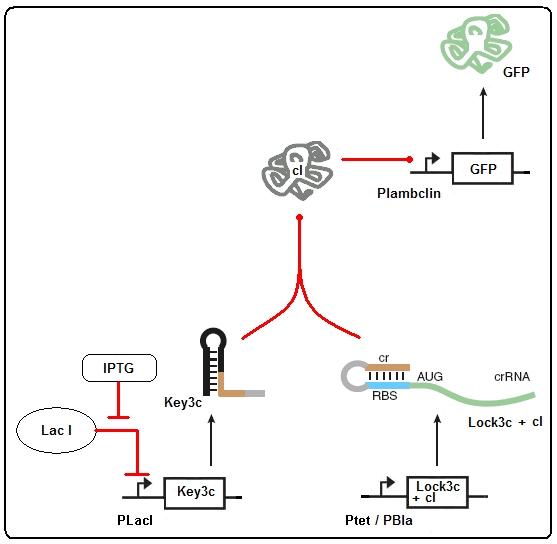
Figure 2: Riboregulator model
Mathematical Modeling
The Negative Cascade model shown above can be expressed with the following set of ODEs (ordinary differential equations):
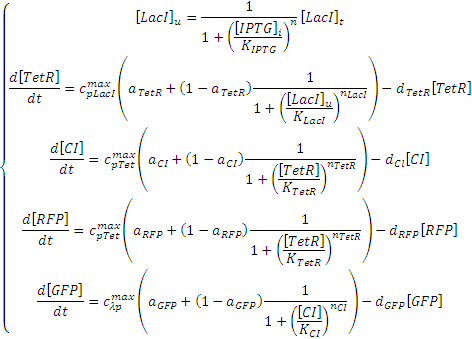
The Riboregulator model shown above can be expressed with the following set of ODEs:
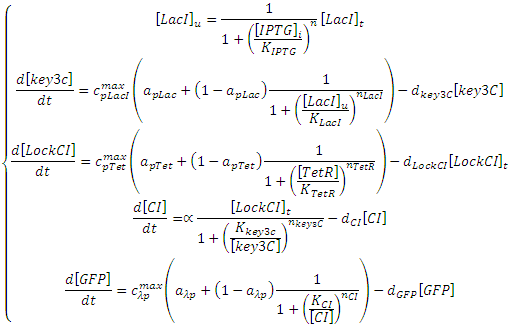
The symbols in this system of equations are found in the table below:
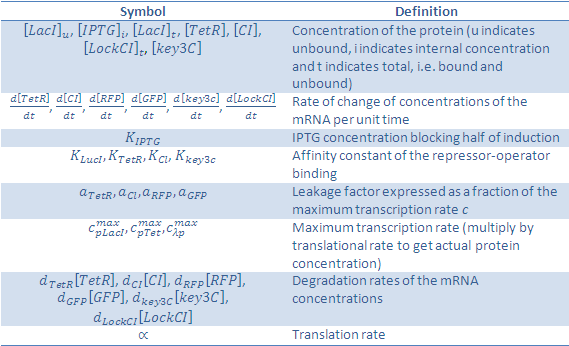
We would like to give credit to the Bologna '08 team and UNIPV-Pavia '08 team, who's project was useful in determining our own equations.
Conjugation
Using the paper "[http://www3.interscience.wiley.com/journal/118837812/abstract?CRETRY=1&SRETRY=0 A model for bacterial conjugal gene transfer on solid surfaces]", we established the assumptions and parameters that would be needed for the modeling of the conjugation.
Assumptions
- Cells are distributed randomly on the agar when introduced to it
- Number of cells grows until one or more of the medium components are exhausted, unless the initial nutrient concentration is lager than the saturation constant for growth (i.e. the ability of the colony to expand)
- Donors, recipients andf transconjugants have identical colony growth rates, specific growth rates and cell yields on solidified LB
- Conjugation occurs such that all recipients become transconjugants after a certain conjugation time
- Plasmid loss is negligable
- Cells only move on the surface through colony expansion
Parameters
- Surface area of media (A)
- Initial colony radius (r_0)
- Specific growth rate (g_n)
- Colony radial growth rate (g_r)
- Maximum numbers of cells sustained by system (N_max)
- Initial number of donors (N_d) and recipients (N_r)
All these parameters can be determined experimentally, or by literary research into previous conjugation experiments.
Spatial distribution of bacteria
The bacteria are initially distributed at random on the medium, so a distribution function to find the closest cell r would be:
F(r)=1-exp(-λ*r^2), where λ is the intensity (the expected number of bacteria per unit area).
Growth of colonies
With g_r being the exponential increasing rate of the colony radius at a certain time period, we can express the radius of the colony at time t as rad(t)=r_0*exp(g_r*t), where r_0 is the initial radius.
With g_n being the exponential increasing rate of the cell number within a colony at a certain time period, we can express the number of bacteria in a colony at time t as N(t)=exp(g_n*t).
Finally, there is a pre-given maximum yield of cells N_max, and growth stops when the total number of cells reaches this number.
Contact between colonies
Two colonies with radius r will touch if their centres are less then 2r apart (i.e. overlapping colonies). Therefore, we can express the probability that the colonies will not be interacting by the function
p(Not touching)=p(radius1+radius2>2r)=p1=exp(-4*r^2*π*λ)
When this is considered as a function of time, the function would be
p1(t)=exp(-4*π*λ*(r_0*exp(g_r*t))^2)=exp(-4*π*λ*(r_0)^2*exp(2*g_r*t))<br
When we approximate the distribution of colony clumps, it depends on the parameter p1, so the probability that a colony chosen at random from the plate is part of a clump containing n colonies is
p_n=p1(1-p1)^(n-1)=exp(-4*r^2*π*λ)(1-exp(-4*r^2*π*λ))^(n-1)
This function in terms of time would be expressed as
p_n(t)=exp(-4*π*λ*(r_0)^2*exp(2*g_r*t))(1-exp(-4*π*λ*(r_0)^2*exp(2*g_r*t)))^(n-1)
In this case, the intensity λ=(N_d+N_r)/A, where A is the area of the medium, and N_d and N_r are the number of donors and recipients at t=0, respectively.
Conjugation modeling
In order for conjugation to take place, the donor and recipient colonies must interact with each other and be in the same clump (depnds on parameter p_n). For a colony chosen at random to be a recipient at time t, it must initially be a recipient and within a clump of only recipient colonies. The probability that any bacterium is a recipient is N_r/(N_d+N_r), and the probability that a clump with n colonies has only recipients is (N_r/(N_d+N_r))^n. To find whether a colony is sampled and a recipient, its probability can be expressed as
p_r=p1*N_r/(N_d+p1*N_r)
Final cell number
The assumptions state that cell numbers increase and conjugation can occur until either the end of the incubation period, t_max, or until all the nutrients have been fully utilised, whichever occurs first. This is modelled by continuing calculations until N_max cells are produced or t = t_max. This time, t_eff , is the minimum of t_max and {ln(N_max)-ln(N_d+N_r)}/gn. Numbers of donor (M_d), recipient (M_r), and transconjugant cells (M_t) at t_max are given
by:
M_d = N_d*exp(g_n*t_eff)
M_r = (N_d+N_r)*p1*N_r*exp(g_n*t_eff)/(N_d+p1*N_r)
M_t = N_r*exp(g_n*t_eff)-M_r
 "
"







