The Registry of Standard Biological Parts is a library of DNA sequences combined with online characterization resources. The Registry has created a standard protocol for making segments of DNA compatible with all other segments of DNA, regardless of order or size. BioBrick is the term given to such segments of DNA, a term alluding to the fact that any number of bricks may be combined in any order to produce complex, unique systems. This is accomplished by standardizing the restriction enzymes used to surround BioBricks, as well as the plasmids used to transform them. For a graphical representation of of the process, please click [http://ginkgobioworks.com/support/BioBrick_Assembly_Manual.jpg here]. A powerful online database provides information and characterization of all of the BioBricks in the Registry and uses the wiki format (the same one used in wikipedia) which encourages others to edit content directly on the page. Below are list of parts that were used/created for this project.
Parts used to characterize and build our final project
| Component | Part/Accession # | Base Pairs | Plasmid | Resistance | Well |
| RBS-34 | [http://partsregistry.org/Part:BBa_B0034 BBa_B0034] | 12 | pSB1A2 | Ampicillin | plate 1, 2M |
| Cph8 | [http://partsregistry.org/Part:BBa_I15010 BBa_I15010] | 2,238 | pSB2K3 | Kanamycin | N/A |
| RFP | [http://partsregistry.org/Part:BBa_J04051 BBa_J04051] | 720 | N/A | N/A | N/A |
| OmpR (E. coli) | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K098011 BBa_K098011] | 720 | pSB1T3 | Tetracycline | N/A |
| OmpR (R. sphaeroides) | [http://partsregistry.org/Part:BBa_K227010 BBa_K227010] | 720 | N/A | N/A | N/A |
| Terminator | [http://partsregistry.org/Part:BBa_B0015 BBa_B0015] | 129 | pSB1AK3 | Ampicillin and Kanamycin | plate 1, 23L |
| RBS +OmpR(sph) + Terminator
includes prefix and suffix | sequence | 916 | pany-amp | Ampicillin | synthesized |
| OmpC promoter | [http://partsregistry.org/Part:BBa_R0082 BBa_R0082] | 108 | pSB1A2 | Ampicillin | plate 1, 16K |
| puc promoter | [http://partsregistry.org/Part:BBa_K227007 BBa_K227007] | 651 | pSB1k3 | Kanamycin | N/A |
| puc BA | [http://partsregistry.org/Part:BBa_K227006 BBa_K227006] | 375 | pSB1k3 | Kanamycin | N/A |
| puc B | [http://partsregistry.org/Part:BBa_K227005 BBa_K227005] | 156 | pSB1k3 | Kanamycin | N/A |
| puc A | [http://partsregistry.org/Part:BBa_K227004 BBa_K227004] | 165 | pSB1k3 | Kanamycin | N/A |
| OmpC promoter+BA | sequence | 539 | pany-kana | Kanamycin | synthesized |
| TetR repressible | [http://partsregistry.org/Part:BBa_J13002 BBa_J13002] | 74 | pSB1A2 | Ampicillin | plate 1, 13B |
| Green Fluorescent Protein | [http://partsregistry.org/Part:BBa_E0240 BBa_E0240] | 876 | pSB1A2 | Ampicillin | plate 1, 12M |
Plasmids used to create and characterize our project
| Plasmid | Base Pairs | Resistance | Copy Number |
| [http://partsregistry.org/Part:pSB1A2 pSB1A2] | 2,079 | Ampicillin | high |
| [http://partsregistry.org/Part:pSB1K3 pSB1K3] | 2,206 | Kanamycin | high |
| [http://partsregistry.org/Part:pSB1A3 pSB1A3] | 2,157 | Ampicillin | high |
| [http://partsregistry.org/Part:pSB2K3 pSB2K3] | 4,425 | Kanamycin | variable |
| [http://partsregistry.org/Part:pSB1T3 pSB1T3] | 2,463 | Tetracycline | high |
| [http://partsregistry.org/Part:pSB1AK3 pSB1AK3] | 3,189 | Ampicillin and Kanamycin | high |
| pANY | |||
| pRK404 | |||
| pRKPLHT7 | Tetracycline | ||
| pRKCBC3 | Tetracycline |
Parts submitted to the Registry of Standard Biological Parts
| Part/Accession #Component | Component | Type | Base Pairs | Plasmid | Resistance |
| [http://partsregistry.org/Part:BBa_I15010 BBa_I15010] | Cph8 (resubmission) | Coding | 2,238 | plasmid | Resistance |
| [http://partsregistry.org/Part:BBa_K227004 BBa_K227004] | puc A | Coding | 165 | pSB1K3 | Kanamycin |
| [http://partsregistry.org/Part:BBa_K227005 BBa_K227005] | puc B | Coding | 156 | pSB1K3 | Kanamycin |
| [http://partsregistry.org/Part:BBa_K227006 BBa_K227006] | puc BA | Coding | 376 | pSB1K3 | Kanamycin |
| [http://partsregistry.org/Part:BBa_K227007 BBa_K227007] | puc promoter | Regulatory | 651 | pSB1K3 | Kanamycin |
| [http://partsregistry.org/Part:BBa_K227008 BBa_K227008] | ompC+PucBA (synthesized) | Composite | 539 | pSB1AT3 | Ampicilin,Tetracycline |
| [http://partsregistry.org/Part:BBa_K227009 BBa_K227009] | PucPromotor+GFP | Composite | 1377 | pSB1A2 | Ampicilin |
| [http://partsregistry.org/Part:BBa_K227011 BBa_K227011] | RBS34+OmpR+Term (synthesized) | Composite | 916? | pSB1K3? | Kanamycin? |
| [http://partsregistry.org/Part:BBa_K227012 BBa_K227012] | RBS34+OmpR(sph)+Term+OmpC+PucB/A | Composite | ??? | pSB1K3 | Kanamycin |
| [http://partsregistry.org/Part:BBa_K227013 BBa_K227013] | ompC + GFP | Composite | 992 | plasmid | Resistance |
| [http://partsregistry.org/Part:BBa_K227014 BBa_K227014] | pucpro+pucBA | Composite | ??? | pSB1K3 | Kanamycin |
| [http://partsregistry.org/Part:BBa_K227015 BBa_K227015] | RBS34+OmpR(sph)+Term+OmpC+GFP | Composite | ??? | plasmid | Ampicilin |
pucB/A
pucB/A are the genes that code for the beta and alpha subunits of the light harvesting complex LH2. Here, we measured the expression level of pucB/A from the puc promoter in the genome vs. on a low copy plasmid, pRKCBC3. This data allows us to utilize pucB/A as a reporter gene for expression from promoters, in complement to its role in cellular growth.
Method
Relative LH2 expression in R. sphaeroides 2.4.1, R. sphaeroides DBCΩ and R. sphaeroides DBCΩ pRKCBC3 grown anaerobically in the dark
Growth conditions: -Cultures grown in the dark at 34° C, shaking at 160 rpm - R. sphaeroides 2.4.1 -15 ml culture in polystyrene tube -15ml M22 was inoculated with 1ml inoculant with OD600nm = 0.270 - OD600nm (Volume) = 0.270 -Culture was allowed to grow 19hrs - R. sphaeroides DBCΩ -15 ml culture in polystyrene tube -15ml M22 was inoculated with 865ul inoculant with OD600nm = 0.312 - OD600nm (Volume) = 0.270 -Culture was allowed to grow 19hrs - R. sphaeroides DBCΩ pRKCBC3 -10 ml culture in polystyrene tube -10 ml M22 tet 5ug/ml was inoculated with loop of R. sphaeroides DBCΩ pRKCBC3 and capped with rubber stopper -Culture was allowed to grow for 3 days so that density was high enough to get a good reading
Analysis:
Cultures were removed from the incubtor and placed on ice to slow changes in cellular composition. 1 ml was extracted from each culture and a UV-vis absorption spectra of the culture was taken from 300-950 nm.
The optical density of the cultures at 600nm was used to normalize the absorption spectrum by division by this value. Background subtraction of spectrophotometer data was performed in Origin 6.1 Software. A ten-point baseline was created by a "positive peak" algorithm then modified to approximate the scattering curve that falls as the inverse fourth power of wavelength.
Results
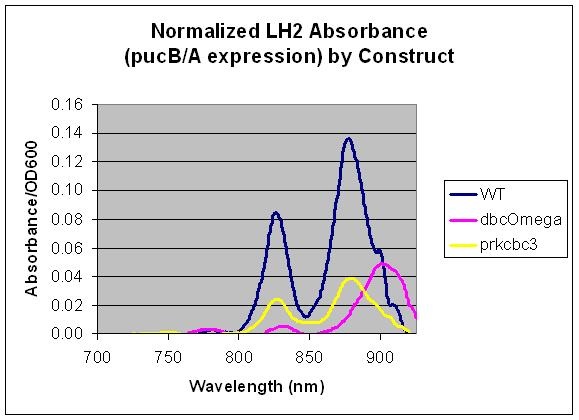
Conclusion
This data indicates that pucB/A can be utilized as a reporter gene as the LH2 absorption bands at 800 and 850 nm are not present in the LH2 deficient mutant DBComega. Furthermore, changes in the expression conditions of pucB/A are reflected on the absorption spectrum. In this case, expression is higher when the genes are expressed from the puc promoter within genomic DNA than on plasmid pRKCBC3, despite the fact that pRKCBC3 is maintained at 4-5 copies in a cell.
Puc Promoter
More absorption of light at the LH2 spectra peaks normalized to culture OD corresponds with more transcription and vis versa.
Method
Cultures were grown in the dark at 34° C, shaking at 160 rpm. The anaerobic test condition was established by inoculating a 10 ml culture tube with 10 ml M22 tet 5ug/ml with a loop of R. sphaeroides DBCΩ pRKCBC3 and capped with a rubber stopper. The aerobic test condition was established by innoculating a 10 ml culture tube with 5ml M22 tet 5ug/ml with a loop of R. sphaeroides DBCΩ pRKCBC3 and covered with vented cap- the vented cap and relatively low culture volume left significant headroom in the culture for oxygen exchange. Oxygen tension was not able to be quantitatively measured due to the nature of the experiment
For measurement of pucB/A expression from the puc promoter:
Cultures were removed from the incubtor and placed on ice to slow changes in cellular composition. 1 ml was extracted from each culture and a UV-vis absorption spectra of the culture was taken from 300-950 nm.
The optical density of the cultures at 600nm was used to normalize the absorption spectrum by division by this value. Background subtraction of spectrophotometer data was performed in Origin 6.1 Software. A ten-point baseline was created by a "positive peak" algorithm then modified to approximate the scattering curve that falls as the inverse fourth power of wavelength.
Results
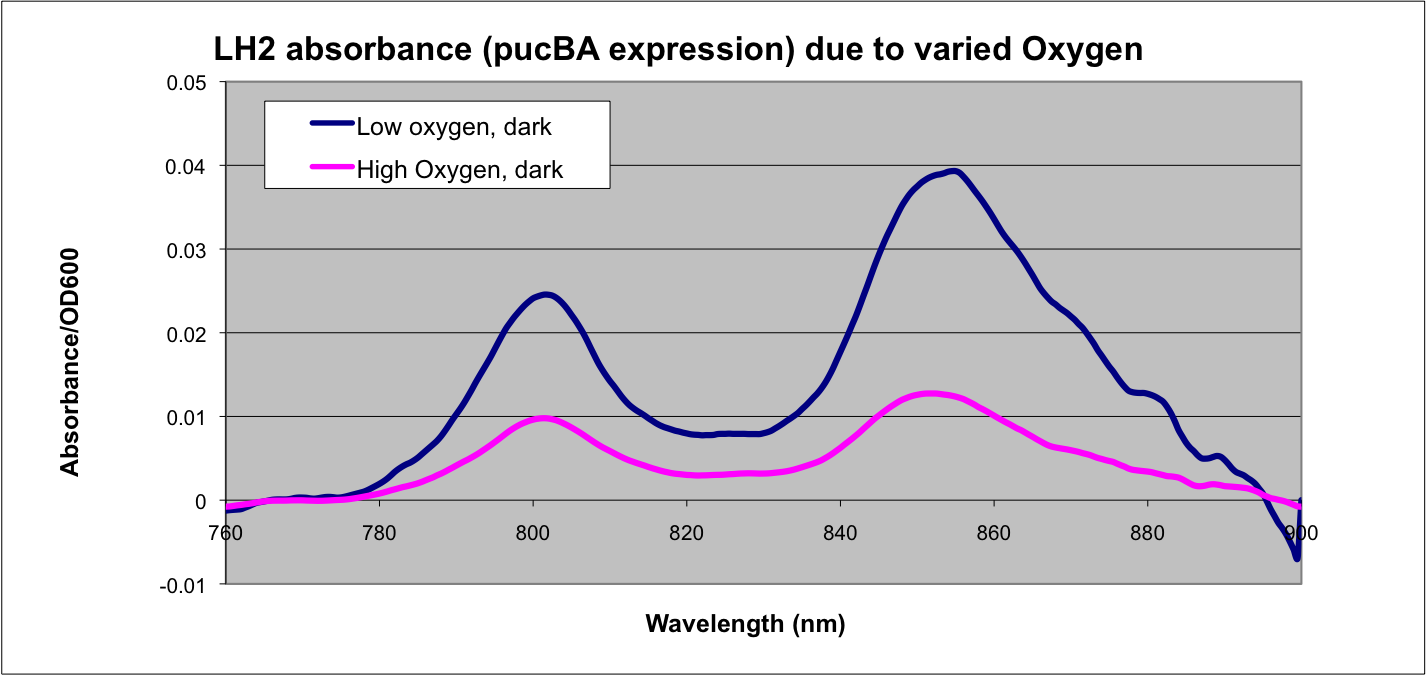
Conclusion
This data matches the literature for expression from the puc promoter at different oxygen tensions and as such confirms the assumptions that we have made in modeling our system.
See: Braatsch et al. 2002
Modeling the Gene Regulatory Network
References
1. Alon, Uri. Introduction to systems biology and the design principles of biological networks. Boca Raton, FL: Chapman & Hall, 2006.
2. Bower, James M. Computational Modeling of Genetic and Biochemical Networks (Computational Molecular Biology). New York: M.I.T. PRESS, 2001.
3. System modeling in cellular biology from concepts to nuts and bolts. Cambridge, MA: MIT P, 2006.
Problem: In a typical reactor, cells at the surface absorb more than enough light to saturate their photosynthetic apparatus, transmitting less energy to deeper layers. For wild type cells, the “saturation curve” is approximately the same for all cells, regardless of their incident light intensity.
Simulating our Mutants advantage in a Bioreactor
For our mutant cells, this curve scales as a function of light intensity, due to negative regulation of LH2 complex production
- Light intensity at next layer is given by transmittance from previous layer (assume no backscattering).
- Total energy funneled to photosynthetic pathways estimated sum of light absorbed by each layer.
type here1
type here2
-Saturation curve: Absorbance as a function of incident light intensity. The coefficient changes with intensity in the mutant only.

