Team:Wash U/Protocol
From 2009.igem.org
(→Polymerase Chain Reaction (PCR)) |
(→Tissue Flask Experiment) |
||
| (62 intermediate revisions not shown) | |||
| Line 18: | Line 18: | ||
# [[Team:Wash_U/Protocol#Gel Extraction|Gel Extraction]] | # [[Team:Wash_U/Protocol#Gel Extraction|Gel Extraction]] | ||
# [[Team:Wash_U/Protocol#Making Competent Cells|Making Competent Cells]] | # [[Team:Wash_U/Protocol#Making Competent Cells|Making Competent Cells]] | ||
| + | # [[Team:Wash_U/Protocol#Tissue Flask Experiment|Tissue Flask Experiment]] | ||
| + | # [[Team:Wash_U/Protocol#R. sphaeroides Protocols and Recipes|R. shpaeroides Protocols and Recipes]] | ||
<font size="4"> | <font size="4"> | ||
| Line 26: | Line 28: | ||
* 5 uL AccuTaq LA 10X Buffer | * 5 uL AccuTaq LA 10X Buffer | ||
* 2.5 uL dNTP mix | * 2.5 uL dNTP mix | ||
| - | * | + | * x uL Template DNA (about 40ng/uL) |
* 1 uL DMSO | * 1 uL DMSO | ||
* 1 uL Forward primer | * 1 uL Forward primer | ||
* 1 uL Reverse Primer | * 1 uL Reverse Primer | ||
| - | |||
* 0.5 uL AccuTaq LA DNA Polymerase | * 0.5 uL AccuTaq LA DNA Polymerase | ||
| + | * dI H20 to 50 uL | ||
'''Procedure''' | '''Procedure''' | ||
# Begin by combining the top 8 ingredients in a microcentrifuge tube-add water until the volume reaches 50 uL. | # Begin by combining the top 8 ingredients in a microcentrifuge tube-add water until the volume reaches 50 uL. | ||
| Line 134: | Line 136: | ||
'''Materials''' | '''Materials''' | ||
* NEBuffer 2 | * NEBuffer 2 | ||
| - | * BSA | + | * 10x BSA |
* dI H20 | * dI H20 | ||
* Upstream, Downstream, and Destination Plasmid parts | * Upstream, Downstream, and Destination Plasmid parts | ||
| Line 140: | Line 142: | ||
'''Procedures''' | '''Procedures''' | ||
# Begin by thawing the upstream, downstream, and destination plasmid parts along with the NEBuffer 2 and BSA. | # Begin by thawing the upstream, downstream, and destination plasmid parts along with the NEBuffer 2 and BSA. | ||
| - | # In three separate PCR microcentrifuge tubes labeled upstream, downstream, and destination, add | + | # In three separate PCR microcentrifuge tubes labeled upstream, downstream, and destination, add 750ng-1000ng of the respective dried DNA and dilute with dH20 to 38 uL. |
| - | # Add 5 uL of NEBuffer 2 and | + | # Add 5 uL of NEBuffer 2 and 5 uL of 10x BSA to each tube. |
# Add 1 uL of the first appropriate enzyme to each tube. Then add 1 uL of the second appropriate enzyme. | # Add 1 uL of the first appropriate enzyme to each tube. Then add 1 uL of the second appropriate enzyme. | ||
| - | # Flick each tube to mix reagents and incubate at 37C for | + | # Flick each tube to mix reagents and incubate at 37C for 1 hour. |
# Transfer the tubes to an incubator set at 80C for another 20 minutes. This step will deactivate the restriction enzymes. | # Transfer the tubes to an incubator set at 80C for another 20 minutes. This step will deactivate the restriction enzymes. | ||
# Digestion is now finished and products should be stored at -20C or proceed to Ligation. | # Digestion is now finished and products should be stored at -20C or proceed to Ligation. | ||
| Line 150: | Line 152: | ||
<font size="4"> | <font size="4"> | ||
| + | |||
=='''Ligation'''== | =='''Ligation'''== | ||
<font size="2"> | <font size="2"> | ||
| Line 163: | Line 166: | ||
# Add 2 uL of 10X T4 DNA Ligase Reaction Buffer to the 200 uL PCR tube. | # Add 2 uL of 10X T4 DNA Ligase Reaction Buffer to the 200 uL PCR tube. | ||
# Add 1 uL DNA Ligase to the PCR tube and flick to ensure the contents are mized. | # Add 1 uL DNA Ligase to the PCR tube and flick to ensure the contents are mized. | ||
| - | # Let the mix stand for | + | # Let the mix stand for 1 hour at room temperature before incubating at 80C for 20 minutes (deactivates enzymes). |
# Store the products at -20C until they are needed for a transformation. | # Store the products at -20C until they are needed for a transformation. | ||
[https://2009.igem.org/Team:Wash_U/Protocol Back To Top] | [https://2009.igem.org/Team:Wash_U/Protocol Back To Top] | ||
| Line 169: | Line 172: | ||
<font size="4"> | <font size="4"> | ||
| + | |||
=='''Gel Electrophoresis'''== | =='''Gel Electrophoresis'''== | ||
<font size="2"> | <font size="2"> | ||
| Line 357: | Line 361: | ||
# Centrifuge again at 4C at 3,000g for 10 minutes. Discard the solution and resuspend the cells in 10mL of CCMB80 buffer. Test OD of a mixture of 200uL SOC and 50uL of the resuspended cells. Add chilled CCMB80 to yield a final OD of 1.0-1.5. | # Centrifuge again at 4C at 3,000g for 10 minutes. Discard the solution and resuspend the cells in 10mL of CCMB80 buffer. Test OD of a mixture of 200uL SOC and 50uL of the resuspended cells. Add chilled CCMB80 to yield a final OD of 1.0-1.5. | ||
# Incubate on ice for 20 minutes and then aliquot to 2mL vials and store indefinitely at -80C. | # Incubate on ice for 20 minutes and then aliquot to 2mL vials and store indefinitely at -80C. | ||
| + | [https://2009.igem.org/Team:Wash_U/Protocol Back To Top] | ||
| + | |||
| + | <font size="4"> | ||
| + | =='''Tissue Flask Experiment'''== | ||
| + | <font size="2">This experiment to monitor the growth and spectra of Rhodobacter shaeroides under anaerobic, photosynthetic conditions was performed as follows:<br> | ||
| + | |||
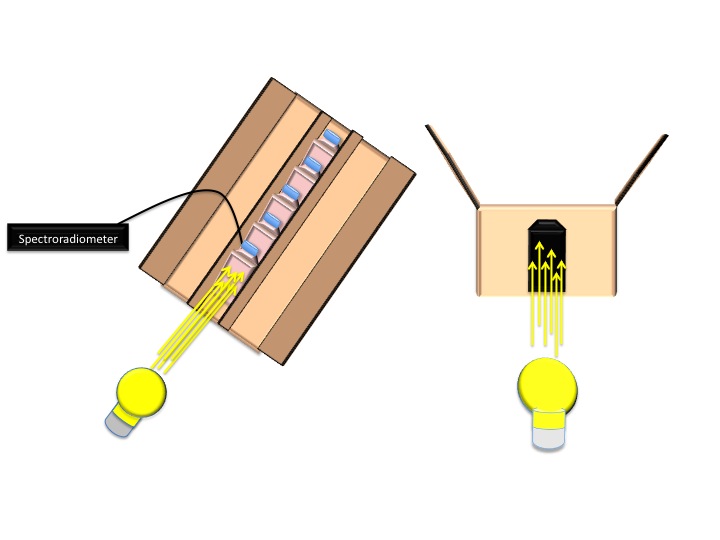
| + | -To ensure optimal growth conditions, an Excella E24 incubator shaker was used to maintain a temperature of 34° C and shaking at 160 rpm and the lid of the incubator was propped open 0.5 cm to prevent overheating. The incubator was covered by a black, opaque cloth to prevent external light from influencing culture growth. Tissue flasks were enclosed by 2 growth boxes inside the incubator, which allowed light to enter only through a tissue flask sized opening on the end nearest the light source. These boxes had the dimensions: 34.8 cm x 18.3cm x 16.8cm.<br> | ||
| + | -5 BD Falcon ™ 50 ml 25 cm² Cell Culture Flasks with blue plug-seal screw caps were used to grow cultures within each box. Cell culture flasks were arranged in series with the first flask recessed 2.5 cm from of box opening and with 2.5 cm separations between each flask. The cell culture flasks were flanked on each side by an opaque barrier so that light incident on each flask was either from the light source, in the case of the 1st flask, or from the light transmittance from the preceding flask in the series<br> | ||
| + | -A constant light source was centered in front of each of the growth boxes The Light source was a 40 watt incandescent light bulb located 10.8 cm from the growth box opening, as measured from the closest point to the box.<br> | ||
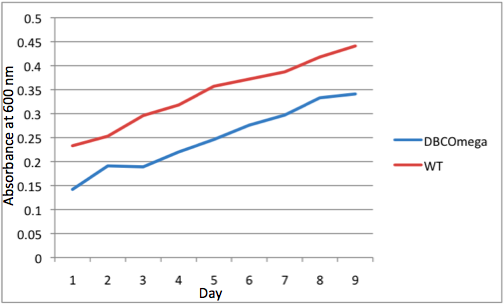
| + | -Tissue culture flasks were inoculated with R. sphaeroides cultures in the exponential growth phase from a 50 ml inoculation culture grown at 34° C in the dark in M22 liquid media. <br> | ||
| + | -The test tissue culture flasks were inoculated with Volume = 0.478ml / OD600nm of inoculation culture <br><br> | ||
| + | [[Image:Innoculation culture growth a.png| 500px | center]]<br> | ||
| + | -M22 liquid media was added to flasks for a final volume of 65ml and just after inoculation each flask had OD600nm = 0.011<br> | ||
| + | -Cell culture growth was monitored by measuring OD600nm daily at a standard time for each tissue culture flask. To conduct this measurement,1ml of culture was extracted from each flask for measurement and 1ml M22 liquid media was added to each flask to replace the extracted volume and to ensure minimal oxygen exposure to the culture by reducing headroom. <br><br> | ||
| + | -Spectroradiometer readings were taken with an Ocean Optics QP-600-1-UV-Vis EOS25354-2 optic cable transmitting data to an Ocean Optics HR2000CG-UV-NIR High-Resolution Spectrometer. Absolute irradiance and transmittance data were recorded in front of the first flask, then behind this flask and each subsequent flask. The absolute irradiance data were calibrated to a Mikropack HL-2000 Halogen Light Source. The transmittance data were recorded relative to the reading at the front of the first flask. | ||
| + | |||
| + | [[Image:tissue flask.jpg| 700px | center]] | ||
| + | |||
| + | [[Image:tissue flask 0981 b.jpg| 450px | left]][[Image:tissue flask 00973.jpg| 450px | right]] | ||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
[https://2009.igem.org/Team:Wash_U/Protocol Back To Top] | [https://2009.igem.org/Team:Wash_U/Protocol Back To Top] | ||
<font size="4"> | <font size="4"> | ||
| + | |||
== '''Recipes''' == | == '''Recipes''' == | ||
<font size="2"> | <font size="2"> | ||
| Line 419: | Line 444: | ||
* sterile filter and store at 4°C | * sterile filter and store at 4°C | ||
* slight dark precipitate appears not to affect its function | * slight dark precipitate appears not to affect its function | ||
| + | |||
| + | [https://2009.igem.org/Team:Wash_U/Protocol Back To Top] | ||
| + | |||
| + | <font size="4"> | ||
| + | |||
| + | == '''''R. sphaeroides'' Protocols and Recipes''' == | ||
| + | '''Optimal Growth Conditions''' | ||
| + | <font size="2"> | ||
| + | *M22 Media, anaerobic | ||
| + | *34º C | ||
| + | *Incandescent Light | ||
| + | |||
| + | '''Recipes''' | ||
| + | <font size="2"><br><br> | ||
| + | '''M22 Growth Media'''<br> | ||
| + | M22 is a Minimal Media that promotes photosynthetic growth. The media is essential for tripaternal mating protocol where it prevents growth of E. coli.<br> | ||
| + | 10X Stock: makes up 4 Liters | ||
| + | *Potassium dihydrogen orthophosphate KH2PO4 122.4g | ||
| + | *Diapotassium dihydrogen orthophosphate K2HPO4 120.0g | ||
| + | *DL – Lactic acid (fridge) Na Lactate solution 100.0g | ||
| + | *Ammonium sulphate – big pot (NH4)2SO4 20g | ||
| + | *Sodium Chloride NaCl 20g | ||
| + | *Sodium succinate 173.7g | ||
| + | *Sodium glutamate L – glutamic acid 10.8g | ||
| + | *Aspartic acid DL – aspartic acid 1.6g | ||
| + | *Solution C 800ml | ||
| + | Make up to 2-3 litres, pH to 6.8 and then make up to 4 Litres. | ||
| + | Autoclave as 10 X 500ml flats. | ||
| + | |||
| + | 1X M22: | ||
| + | To make up 2 Litres | ||
| + | *10X stock M22 200ml | ||
| + | *CAA 40ml | ||
| + | *Water 1760ml | ||
| + | |||
| + | |||
| + | '''Solution C'''<br> | ||
| + | Makes up to 4 Liters | ||
| + | *Nitrilotriacetic acid 40g | ||
| + | *Magnesium Chloride MgCl2 96g | ||
| + | *Calcium Chloride CaCl2 13.36g | ||
| + | *EDTA 0.5g | ||
| + | *Zinc Chloride (poison) ZnCl2 1.044g | ||
| + | *Ferrous Chloride (poison) FeCl2 1.0g | ||
| + | *Manganous Chloride MnCl2 0.36g | ||
| + | *Ammonium molybdate (NH4)6Mo7O244H2O 0.037g | ||
| + | *Cupric Chloride (poison) CuCl2 0.031g | ||
| + | *Cobaltous nitrate (poison) Co(NO3)2 0.0496g | ||
| + | *Boric acid (orthoboric acid) 0.0228g | ||
| + | |||
| + | Do not autoclave, just freeze at -20°C in 400ml aliquots. | ||
| + | |||
| + | Casamino acids (CAA): | ||
| + | To make 1 Litre | ||
| + | *Casein Hydrosylate acid 50g | ||
| + | Makes up 5% solution to be aliquotted into 200ml. | ||
| + | |||
| + | |||
| + | Batches: | ||
| + | *1.5 Liters in 2 Liter flasks | ||
| + | *12 X 80ml in 100ml flasks | ||
| + | *100 X 10ml in universals. | ||
| + | *For M22 agar add 1.5g agar to 100ml of M22 with no CAA in, store in 300ml flats. | ||
| + | |||
| + | |||
| + | '''Vitamin Solution'''<br> | ||
| + | prepare a 10,000 times stock solution of vitamins as follows. | ||
| + | |||
| + | *nicotinic acid 1g | ||
| + | *Thiamine 0.5g | ||
| + | *pABA(p-Aminobenzoic acid) 0.1g | ||
| + | *Biotin (d-Biotin) 0.01g | ||
| + | *Milli-Q water 100ml | ||
| + | |||
| + | Aliquot into 20mls after filter sterilisation. | ||
| + | |||
| + | Add 1ul per every 10ml of M22 media. Do this after you have autoclaved the | ||
| + | media, since the vitamins are labile and the heat will destroy them. When | ||
| + | adding them to melted agar wait until the agar is relatively cool. Freeze the | ||
| + | aliquots you aren't using and keep your working stock in the fridge. | ||
| + | |||
| + | [https://2009.igem.org/Team:Wash_U/Protocol Back To Top] | ||
| + | |||
| + | |||
| + | <font size="4"> | ||
| + | '''Protocols''' | ||
| + | <font size="2"> | ||
| + | |||
| + | |||
| + | Work was done in E. coli and final constructs were transferred to pRKCBC3, which is based on the broad-host range expression plasmid pRK404. This vector contains sequences for conjugal transfer (oriT), replication (oriV). It is maintained at a low copy number (3-4) and features tetracycline resistance (tetR). | ||
| + | |||
| + | '''Conjugation: Triparental Mating''' | ||
| + | *Plasmid (pRK404 derived) in E. coli GC5 (or other F- strain) | ||
| + | *Conjugation helper strain: E. coli with pRK2013 | ||
| + | *Destination strain: R. sphaeroides DBCΩ(LH2 Knockout with streptomycin resistance) | ||
| + | *Expect to see colonies in 4-6 days | ||
| + | |||
| + | 1) Start cultures | ||
| + | *Start 15ml 2 day culture R. sphaeroides DBCΩ with strep 10ug/ml in M22 | ||
| + | *Start 15 ml overnight culture E. coli GC5 with plasmid to transfer in LB with strep 5ug/ml tet 5ug/ml | ||
| + | *Start 15ml overnight culture E. coli with pRK2013 in LB with kan 50ug/ml<br> | ||
| + | 2) Spin down cultures for 5 minutes<br> | ||
| + | 3) Remove supernatant from cultures and resuspend in 1ml antibiotic-free liquid media | ||
| + | *Use LB for E. coli | ||
| + | *Use M22 for R. sphaeroides DBCΩ<br> | ||
| + | 4) Combine equal volumes of resuspended cells (~30ul each)<br> | ||
| + | 5) Pipette mixture of cells onto well dried M22 plate<br> | ||
| + | 6) Allow cells to dry into media for 8hrs at 34º C in the dark<br> | ||
| + | 7) Scrape dried cells of M22 plate and streak onto M22 step 5ug/ml tet 5ug/ml | ||
[https://2009.igem.org/Team:Wash_U/Protocol Back To Top] | [https://2009.igem.org/Team:Wash_U/Protocol Back To Top] | ||
Latest revision as of 03:49, 22 October 2009

 "
"