From 2009.igem.org
(Difference between revisions)
|
|
| Line 44: |
Line 44: |
| | | | |
| | | | |
| - | Samples: | + | Samples: Fusion protein report 1, 2, 3, 4. |
| | + | <br> |
| | + | |
| | + | [[image:W0710.png|center]] |
| | + | <br> |
| | | | |
| | We obtain a fragment at 385bp, so the ligation does not work as expected. The fragment at 385bp is similar to the D protein, so there is not enough adenovirus 5 penton base into the sample or there is no DNA. <br><br> | | We obtain a fragment at 385bp, so the ligation does not work as expected. The fragment at 385bp is similar to the D protein, so there is not enough adenovirus 5 penton base into the sample or there is no DNA. <br><br> |
Revision as of 16:31, 21 October 2009



| August
|
| M | T | W | T | F | S | S
|
|
|
|
|
|
| [http://2009.igem.org/User:DavidC/1_August_2009 1]
| [http://2009.igem.org/User:DavidC/2_August_2009 2]
|
| [http://2009.igem.org/User:DavidC/3_August_2009 3]
| [http://2009.igem.org/User:DavidC/4_August_2009 4]
| [http://2009.igem.org/User:DavidC/5_August_2009 5]
| [http://2009.igem.org/User:DavidC/6_August_2009 6]
| [http://2009.igem.org/User:DavidC/7_August_2009 7]
| [http://2009.igem.org/User:DavidC/8_August_2009 8]
| [http://2009.igem.org/User:DavidC/9_August_2009 9]
|
| [http://2009.igem.org/User:DavidC/10_August_2009 10]
| [http://2009.igem.org/User:DavidC/11_August_2009 11]
| [http://2009.igem.org/User:DavidC/12_August_2009 12]
| [http://2009.igem.org/User:DavidC/13_August_2009 13]
| [http://2009.igem.org/User:DavidC/14_August_2009 14]
| [http://2009.igem.org/User:DavidC/15_August_2009 15]
| [http://2009.igem.org/User:DavidC/16_August_2009 16]
|
| [http://2009.igem.org/User:DavidC/17_August_2009 17]
| [http://2009.igem.org/User:DavidC/18_August_2009 18]
| [http://2009.igem.org/User:DavidC/19_August_2009 19]
| [http://2009.igem.org/User:DavidC/20_August_2009 20]
| [http://2009.igem.org/User:DavidC/21_August_2009 21]
| [http://2009.igem.org/User:DavidC/22_August_2009 22]
| [http://2009.igem.org/User:DavidC/23_August_2009 23]
|
| [http://2009.igem.org/User:DavidC/24_August_2009 24]
| [http://2009.igem.org/User:DavidC/25_August_2009 25]
| [http://2009.igem.org/User:DavidC/26_August_2009 26]
| [http://2009.igem.org/User:DavidC/27_August_2009 27]
| [http://2009.igem.org/User:DavidC/28_August_2009 28]
| [http://2009.igem.org/User:DavidC/29_August_2009 29]
| [http://2009.igem.org/User:DavidC/30_August_2009 30]
|
| [http://2009.igem.org/User:DavidC/31_August_2009 31]
|
|
| September
|
| M | T | W | T | F | S | S
|
|
| [http://2009.igem.org/User:DavidC/1_September_2009 1]
| [http://2009.igem.org/User:DavidC/2_September_2009 2]
| [http://2009.igem.org/User:DavidC/3_September_2009 3]
| [http://2009.igem.org/User:DavidC/4_September_2009 4]
| [http://2009.igem.org/User:DavidC/5_September_2009 5]
| [http://2009.igem.org/User:DavidC/6_September_2009 6]
|
| [http://2009.igem.org/User:DavidC/7_September_2009 7]
| [http://2009.igem.org/User:DavidC/8_September_2009 8]
| [http://2009.igem.org/User:DavidC/9_September_2009 9]
| [http://2009.igem.org/User:DavidC/10_September_2009 10]
| [http://2009.igem.org/User:DavidC/11_September_2009 11]
| [http://2009.igem.org/User:DavidC/12_September_2009 12]
| [http://2009.igem.org/User:DavidC/13_September_2009 13]
|
| [http://2009.igem.org/User:DavidC/14_September_2009 14]
| [http://2009.igem.org/User:DavidC/15_September_2009 15]
| [http://2009.igem.org/User:DavidC/16_September_2009 16]
| [http://2009.igem.org/User:DavidC/17_September_2009 17]
| [http://2009.igem.org/User:DavidC/18_September_2009 18]
| [http://2009.igem.org/User:DavidC/19_September_2009 19]
| [http://2009.igem.org/User:DavidC/20_September_2009 20]
|
| [http://2009.igem.org/User:DavidC/21_September_2009 21]
| [http://2009.igem.org/User:DavidC/22_September_2009 22]
| [http://2009.igem.org/User:DavidC/23_September_2009 23]
| [http://2009.igem.org/User:DavidC/24_September_2009 24]
| [http://2009.igem.org/User:DavidC/25_September_2009 25]
| [http://2009.igem.org/User:DavidC/26_September_2009 26]
| [http://2009.igem.org/User:DavidC/27_September_2009 27]
|
| [http://2009.igem.org/User:DavidC/28_September_2009 28]
| [http://2009.igem.org/User:DavidC/29_September_2009 29]
| [http://2009.igem.org/User:DavidC/30_September_2009 30]
|
|
| October
|
| M | T | W | T | F | S | S
|
|
|
|
| [http://2009.igem.org/User:DavidC/1_October_2009 1]
| [http://2009.igem.org/User:DavidC/2_October_2009 2]
| [http://2009.igem.org/User:DavidC/3_October_2009 3]
| [http://2009.igem.org/User:DavidC/4_October_2009 4]
|
| [http://2009.igem.org/User:DavidC/5_October_2009 5]
| [http://2009.igem.org/User:DavidC/6_October_2009 6]
| [http://2009.igem.org/User:DavidC/7_October_2009 7]
| [http://2009.igem.org/User:DavidC/8_October_2009 8]
| [http://2009.igem.org/User:DavidC/9_October_2009 9]
| [http://2009.igem.org/User:DavidC/10_October_2009 10]
| [http://2009.igem.org/User:DavidC/11_October_2009 11]
|
| [http://2009.igem.org/User:DavidC/12_October_2009 12]
| [http://2009.igem.org/User:DavidC/13_October_2009 13]
| [http://2009.igem.org/User:DavidC/14_October_2009 14]
| [http://2009.igem.org/User:DavidC/15_October_2009 15]
| [http://2009.igem.org/User:DavidC/16_October_2009 16]
| [http://2009.igem.org/User:DavidC/17_October_2009 17]
| [http://2009.igem.org/User:DavidC/18_October_2009 18]
|
| [http://2009.igem.org/User:DavidC/19_October_2009 19]
| [http://2009.igem.org/User:DavidC/20_October_2009 20]
| [http://2009.igem.org/User:DavidC/21_October_2009 21]
| [http://2009.igem.org/User:DavidC/22_October_2009 22]
| [http://2009.igem.org/User:DavidC/23_October_2009 23]
| [http://2009.igem.org/User:DavidC/24_October_2009 24]
| [http://2009.igem.org/User:DavidC/25_October_2009 25]
|
| [http://2009.igem.org/User:DavidC/26_October_2009 26]
| [http://2009.igem.org/User:DavidC/27_October_2009 27]
| [http://2009.igem.org/User:DavidC/28_October_2009 28]
| [http://2009.igem.org/User:DavidC/29_October_2009 29]
| [http://2009.igem.org/User:DavidC/30_October_2009 30]
| [http://2009.igem.org/User:DavidC/31_October_2009 31]
|
|
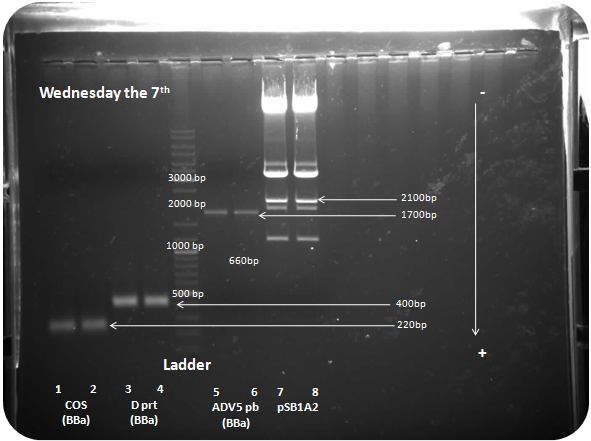
Wednesday the 7th
PCR
Trial for amplification of ligation between D protein and adenovirus 5 penton base
Sample:
D protein (D prt) and adenovirus 5 penton base (ADV5 pb) digest by Spe I and Xba I (samples created on the 2nd of October).
Primers:
Forward : Pref_ATG_Fw
Reverse : Suffixe_Rv
DNA:
5µL of each samples: first, second, third and fourth reports.
Cycles:
94°C 2minutes;
(94°C 1 minute;
59°C 1 minutes;
72°C 2 minute(s)) X 35;
72°C 5 minutes;
DNA electrophoresis
85 Volt, 15 minutes.
105 Volt, 40 minutes.
Ladder fermentas 1 Kb.
Samples: Fusion protein report 1, 2, 3, 4.
We obtain a fragment at 385bp, so the ligation does not work as expected. The fragment at 385bp is similar to the D protein, so there is not enough adenovirus 5 penton base into the sample or there is no DNA.
We will change the next protocol and try to integrate the D protein and the ADV5 pb into plasmid vector independently and join them as a plasmid vector and a insert.
Restriction digest
Restriction digest of adenovirus 5 penton base (1715bp), COS (250bp) and D protein (380bp) by Xba I and Pst I :
DNA (PCR) = 10µL
Buffer M (TAKARA) = 2µL
H20 = 6µL
Xba I (TAKARA) = 1µL
Pst I (TAKARA) = 1µL
1 hour of incubation at 37°C.
Restriction digest of pSB1A2 Xba I and Pst I (2079bp):
DNA (PCR) = 10µL
Buffer M (TAKARA) = 2µL
H20 = 6µL
Xba I (TAKARA) = 1µL
Pst I (TAKARA) = 1µL
1 hour of incubation at 37°C.
DNA electrophoresis
85 Volt, 15 minutes.
105 Volt, 40 minutes.
Ladder fermentas 1 Kb.
Samples: COS with Biobrick prefix and sufix (COS BBa)(250bp), D protein with Biobrick prefix and sufix (D prt BBa)(385bp), adenovirus penton base with Biobrick prefix and sufix (ADV5pb BBa)(1715bp), pSB1A2 (2079bp).
DNA purification
Kit Qiagen “gel extraction kit”, final volume = 50µL.
Ligation
Ligation between ADV5 pb and pSB1A2:
First report:
Plasmid (pSB1A2) = 2µL
Insert (ADV5 pb) = 6µL
Solution A = 1µL
Solution B = 1µL
Second report:
Plasmid (pSB1A2) = 3,5µL
Insert (ADV5 pb) = 10µL
Solution A = 2µL
Solution B = 2µL
2 hours of incubation at room temperature.
Same protocol for:
Ligation of COS with pSB1A2.
Ligation of D protein with pSB1A2.
 "
"